DNA Nanocomputing DEREK WILLINGHAM Overview Introduction to DNA






- Slides: 9

DNA Nanocomputing DEREK WILLINGHAM

Overview • Introduction to DNA nanocomputing • Adleman and the start of DNA nanocomputing • Advances through the 90’s and 2000’s • Current DNA nanocomputing research • Obstacles and future research areas • Summary

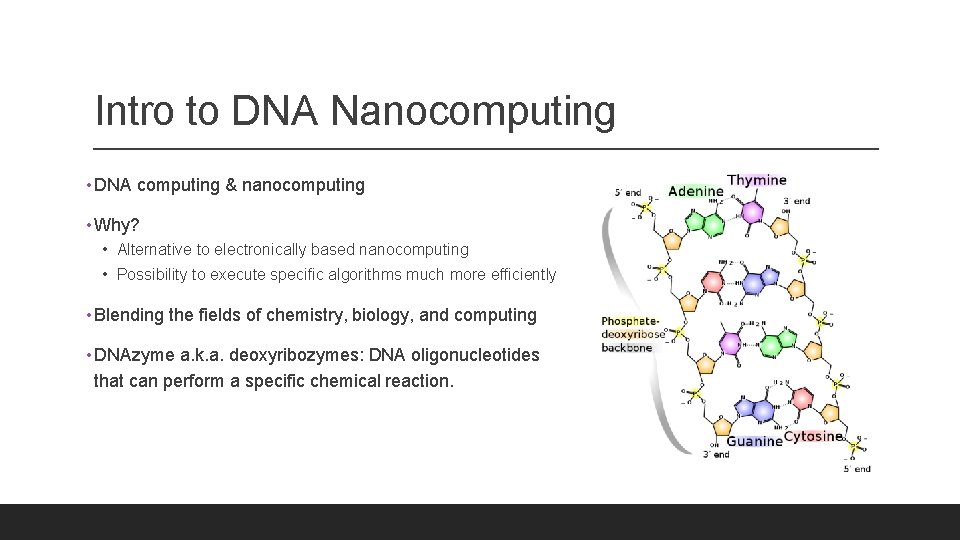
Intro to DNA Nanocomputing • DNA computing & nanocomputing • Why? • Alternative to electronically based nanocomputing • Possibility to execute specific algorithms much more efficiently • Blending the fields of chemistry, biology, and computing • DNAzyme a. k. a. deoxyribozymes: DNA oligonucleotides that can perform a specific chemical reaction.

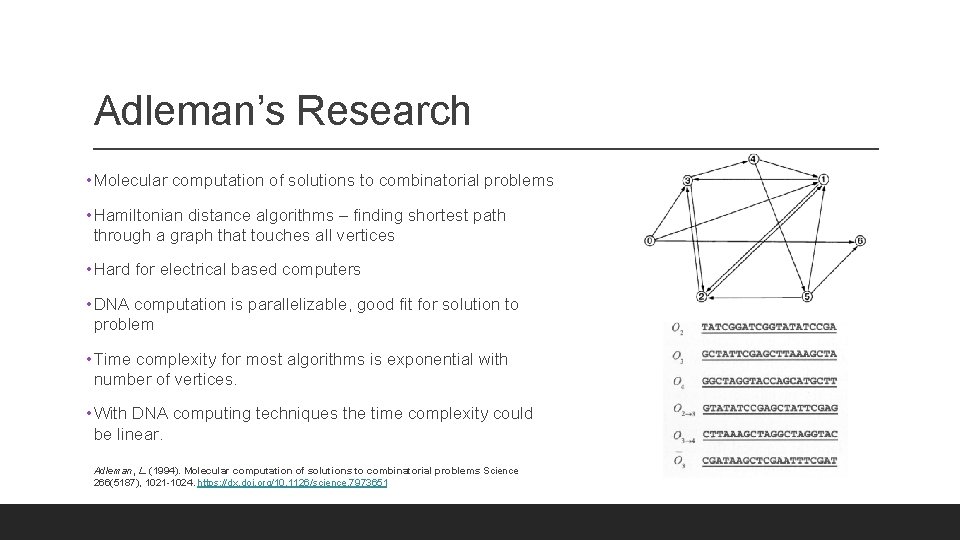
Adleman’s Research • Molecular computation of solutions to combinatorial problems • Hamiltonian distance algorithms – finding shortest path through a graph that touches all vertices • Hard for electrical based computers • DNA computation is parallelizable, good fit for solution to problem • Time complexity for most algorithms is exponential with number of vertices. • With DNA computing techniques the time complexity could be linear. Adleman, L. (1994). Molecular computation of solutions to combinatorial problems Science 266(5187), 1021 -1024. https: //dx. doi. org/10. 1126/science. 7973651

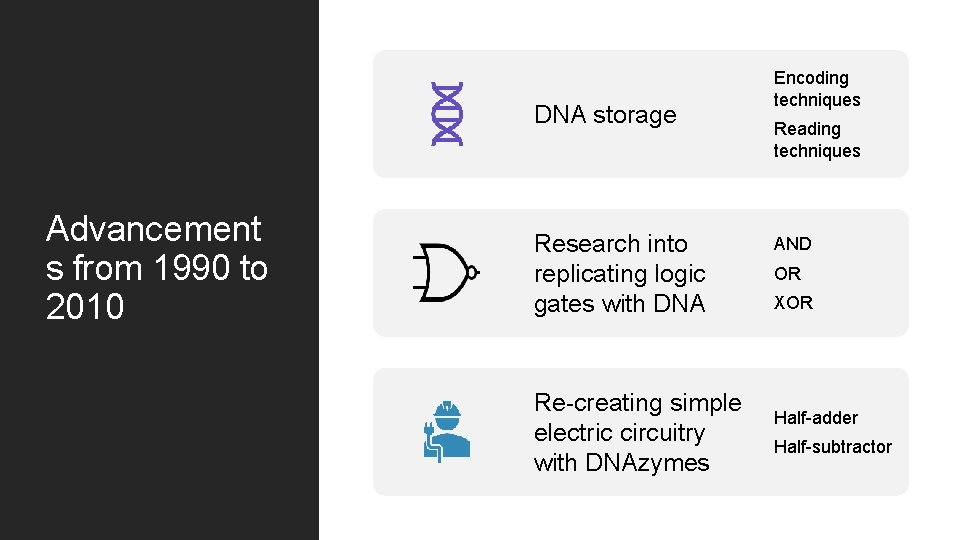
DNA storage Advancement s from 1990 to 2010 Research into replicating logic gates with DNA Re-creating simple electric circuitry with DNAzymes Encoding techniques Reading techniques AND OR XOR Half-adder Half-subtractor

Current DNA Nanocomputing Research • Medical field applications • Improving on previous research, trying to build more complex circuits with DNA strands • Encryption & Decryption possibilities • Finding new, scalable techniques that can bring the benefits of DNA information storage to large scale applications • Recently North Carolina State University researchers invented DENSe, a technology to overcome the 30, 000 -primer sequence limit for DNA strands. • DENSe: DNA Enrichment and Nested Separation • Possible solutions to genetic defects and diseases • Extending life by preventing/repairing DNA degradation https: //phys. org/news/2019 -06 -key-obstacles-scaling-dna-storage. html

Current Research cont. • Most of the experiments carried out use “soup computing”, mixing all ingredients into a solution and the reading the result. • Alternative mode of computation is like how cells use DNA internally • DNA walkers can be used to traverse folded DNA “origami” structures by traveling between anchorages. • Anchorages can be dynamically blocked/unblocked.

Obstacles • Moral and Ethical questions regarding DNA and gene sequence editing • Electric/Organic interface development • Accuracy of calculations • Purity of samples • Cross contamination • DNA is fragile, can break • Most computations in DNA are currently just a sequence of test tubes.

Summary • Many researchers believe advances in the biotechnology field are happening at a similar rate to the silicon industry. • DNA nanocomputing is an extensive field that ranges from utilizing DNA as a storage device to finding ways to compute on DNA strands. • The future is bright for DNA computing and advancements will greatly benefit the computing and medical fields.