DNA FINGERPRINTING INTRODUCTION DNA fingerprinting is a method










- Slides: 35

DNA FINGERPRINTING

INTRODUCTION • DNA fingerprinting is a method of isolating and making images of sequences of DNA. • the technique was developed in 1984 by the British geneticist Alec Jeffreys by RFLP analysis, after he noticed the existence of certain repetitive sequences of DNA (called minisatellites) that do not contribute to the function of a gene. • Jeffreys also determined that each organism has a unique pattern of these minisatellites, the only exception being multiple individuals from a single zygote e. g. , identical twins).

DNA profiles detect and record polymorphisms in the genomes of individuals. DNA profiles provide strong evidence of individual identity, evidence that is extremely valuable in paternity and forensic cases.

RESTRICTION ENZYMES • Restriction enzymes cut DNA but only at a certain combination of A, G, T, and C. • Different restriction enzymes cut DNA at different places—each has a unique sequence it recognizes. • Ex. The restriction enzyme Eco. RI cuts DNA at the sequence GAATTC and will cut only at that sequence.

RFLPs RESTRICTION FRAGMENT LENGTH POLYMORPHISM. • RFLPs are different fragment lengths of base pairs that result from cutting a DNA molecule with a restriction enzyme. • It is the length differences associated with DNA strands or RFLPs that allow one to distinguish one person from another.


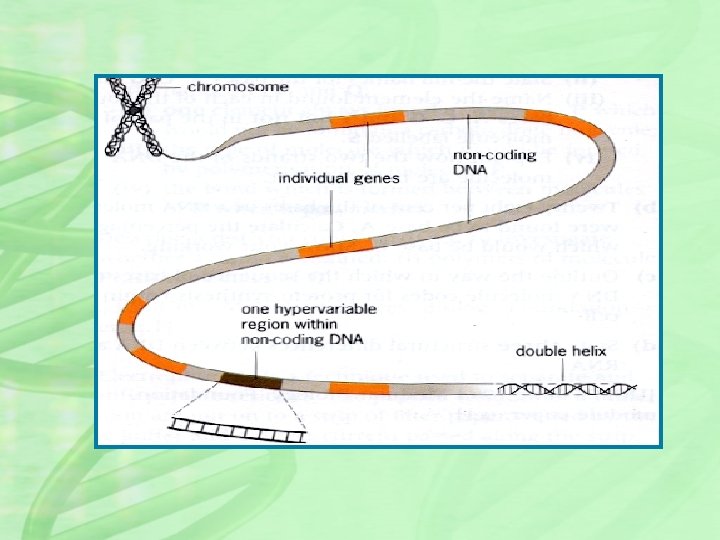
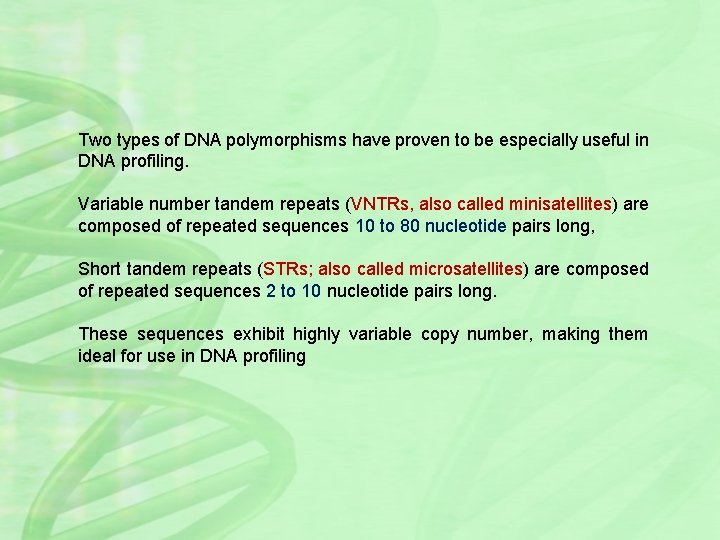
Two types of DNA polymorphisms have proven to be especially useful in DNA profiling. Variable number tandem repeats (VNTRs, also called minisatellites) are composed of repeated sequences 10 to 80 nucleotide pairs long, Short tandem repeats (STRs; also called microsatellites) are composed of repeated sequences 2 to 10 nucleotide pairs long. These sequences exhibit highly variable copy number, making them ideal for use in DNA profiling

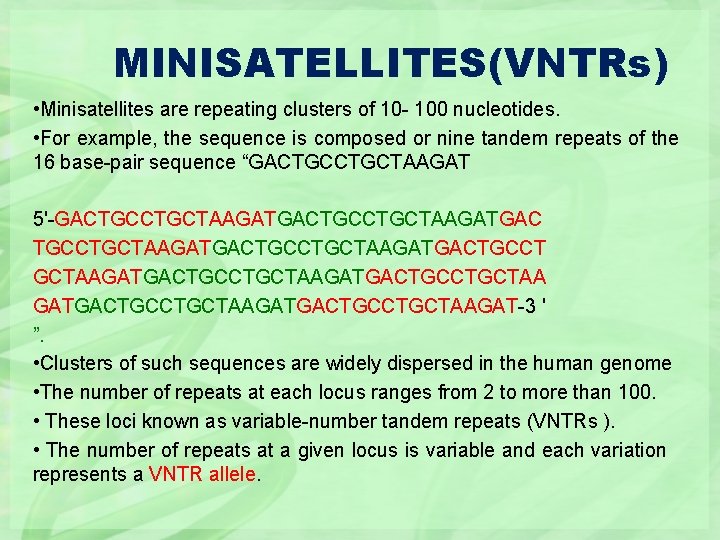
MINISATELLITES(VNTRs) • Minisatellites are repeating clusters of 10 - 100 nucleotides. • For example, the sequence is composed or nine tandem repeats of the 16 base-pair sequence “GACTGCTAAGAT 5'-GACTGCCTGCTAAGATGACTGCCT GCTAAGATGACTGCCTGCTAAGAT-3 ' ”. • Clusters of such sequences are widely dispersed in the human genome • The number of repeats at each locus ranges from 2 to more than 100. • These loci known as variable-number tandem repeats (VNTRs ). • The number of repeats at a given locus is variable and each variation represents a VNTR allele.

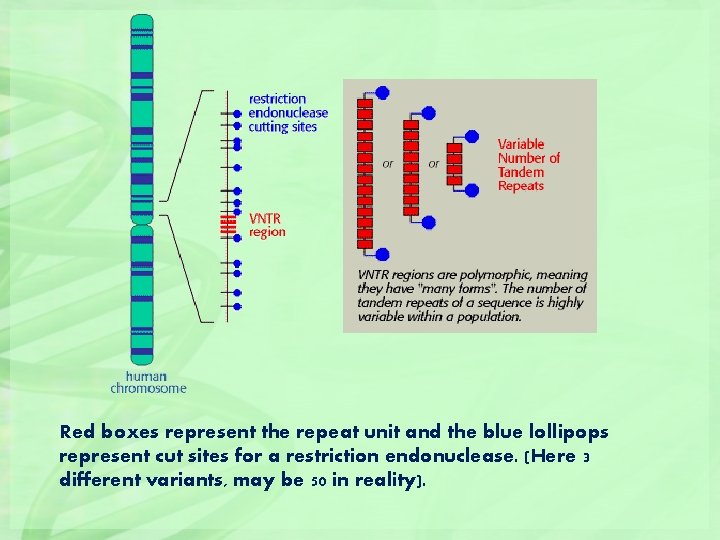
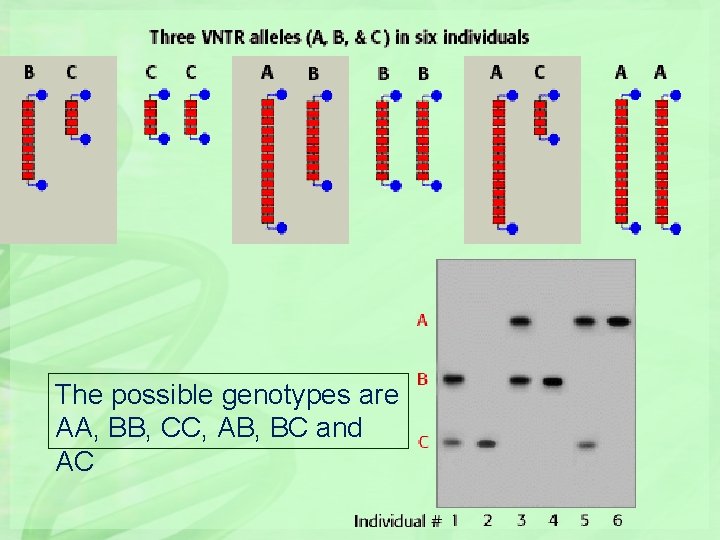
Red boxes represent the repeat unit and the blue lollipops represent cut sites for a restriction endonuclease. (Here 3 different variants, may be 50 in reality).

The possible genotypes are AA, BB, CC, AB, BC and AC

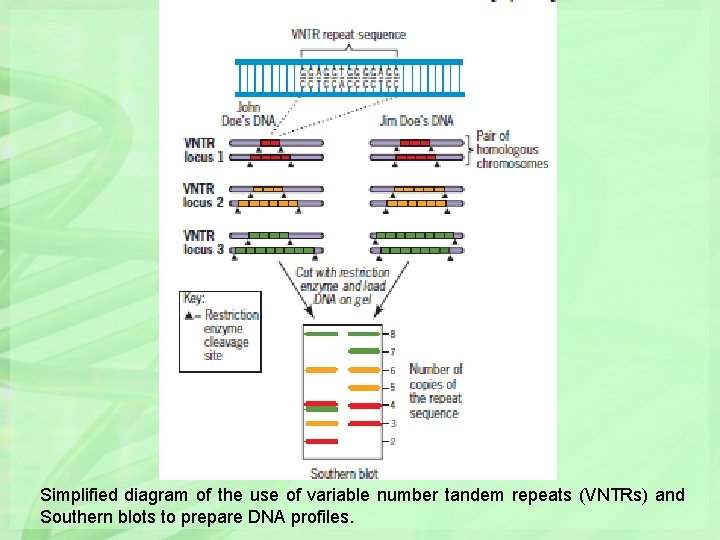
Simplified diagram of the use of variable number tandem repeats (VNTRs) and Southern blots to prepare DNA profiles.

LIMITATIONS OF VNTRs • Limitation of DNA fingerprint analysis with VNTRs is that it requires a relatively large sample of DNA ( 10, 000 cells or about 50 microgram) - more than is usually found at a typical crime scene and the DNA must be relatively intact (no degraded). As a result, DNA fingerprinting is more useful in paternity testing than in criminal cases. In paternity testing. blood drawn from the child, mother and alleged father provide an unlimited source of fresh intact cells for DNA extraction and analysis.

MICROSATELLITES(SRTs) • To overcome the problems in DNA sample size and conditions necessary for analysis with VNTRs a different but related set of sequences are used as markers and are analyzed by PCR rather than RFLP analysis. These sequences called short tandem repeats(STRs). • They are very similar to VNTRs but the repeated motif is shorter between 2 and 9 base pairs. • 13 tetrameric (4 base-pair repeat) STRs have been developed into a marker panel (called the Combined DNA Index System or CODIS panel) used by the FBI • STR typing is less expensive , less labour-intensive and much faster than RFLP analysis

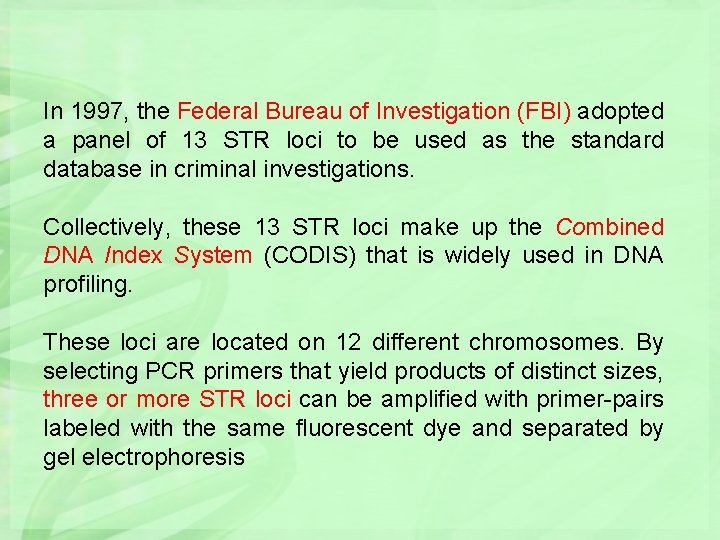
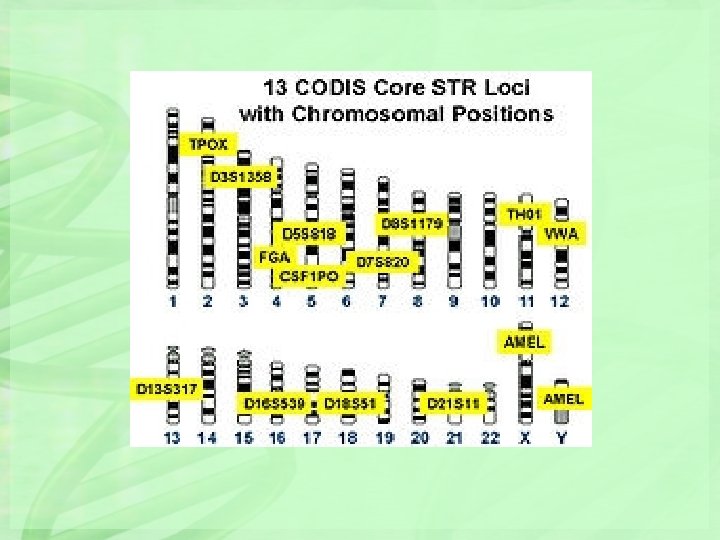
In 1997, the Federal Bureau of Investigation (FBI) adopted a panel of 13 STR loci to be used as the standard database in criminal investigations. Collectively, these 13 STR loci make up the Combined DNA Index System (CODIS) that is widely used in DNA profiling. These loci are located on 12 different chromosomes. By selecting PCR primers that yield products of distinct sizes, three or more STR loci can be amplified with primer-pairs labeled with the same fluorescent dye and separated by gel electrophoresis

The 13 STR Loci in the Core CODIS Panel Locus 1. TPOX 2. D 3 S 1358 3. FGA 4. D 5 S 818 5. CSF 1 PO 6. D 7 S 820 7. D 8 S 1179 8. TH 01 9. VWA 10. D 13 S 317 11. D 16 S 539 12. D 18 S 51 13. D 21 S 11 Chromosome 2 3 4 5 5 7 8 11 12 13 16 18 21 Repeat Motif GAAT [TCTG][TCTA] CTTT AGAT TAGA GATA [TCTA][TCTG] TCAT [TCTG][TCTA] TATC GATA AGAA [TCTA][TCTG] Number of Alleles Observed 15 25 80 15 20 30 15 20 29 17 19 51 89


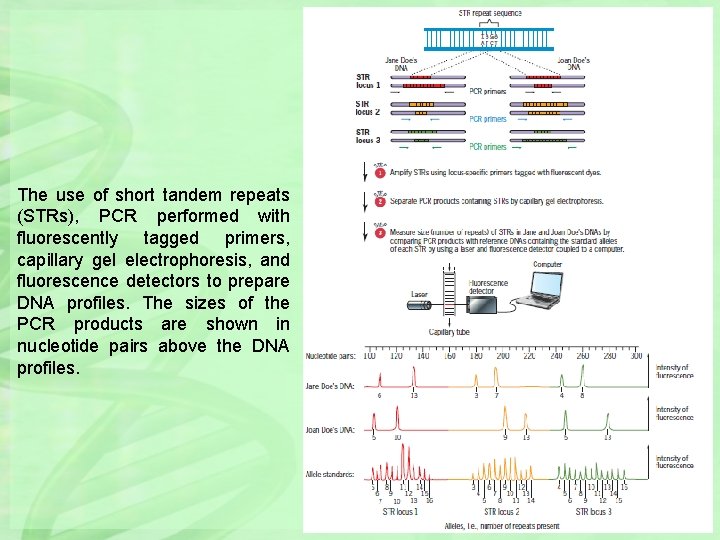
The use of short tandem repeats (STRs), PCR performed with fluorescently tagged primers, capillary gel electrophoresis, and fluorescence detectors to prepare DNA profiles. The sizes of the PCR products are shown in nucleotide pairs above the DNA profiles.

Up to nine STR loci can be amplified using three PCR primer-pairs labeled with distinct fluorescent dyes and separated in a single capillary gel electrophoresis tube. The separation of families of STR alleles in one to three PCR amplifications and one or two gel electrophoresis separations is called multiplex STR analysis. Several companies have developed fluorescent dye-labeled multiplex PCR primers that allow characterization of the alleles of all 13 standard STR loci in just two PCR amplifications and gel electrophoresis separations.

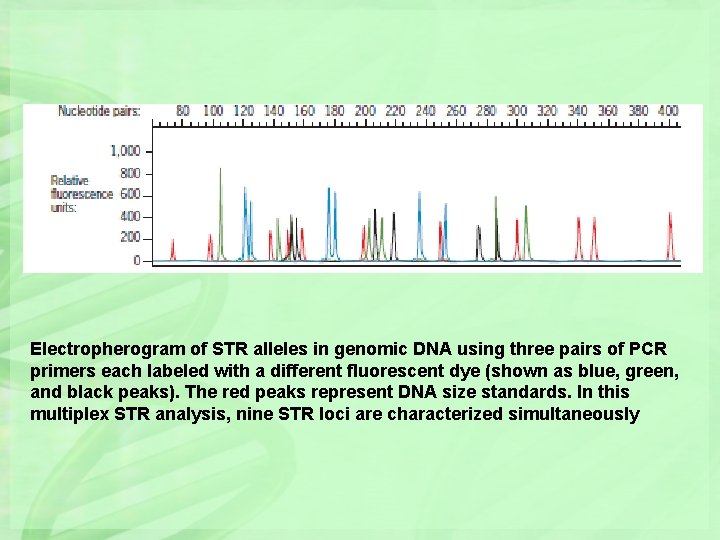
Electropherogram of STR alleles in genomic DNA using three pairs of PCR primers each labeled with a different fluorescent dye (shown as blue, green, and black peaks). The red peaks represent DNA size standards. In this multiplex STR analysis, nine STR loci are characterized simultaneously

Biological materials used for DNA profiling • Blood • Hair • Saliva • Semen • Body tissue cells

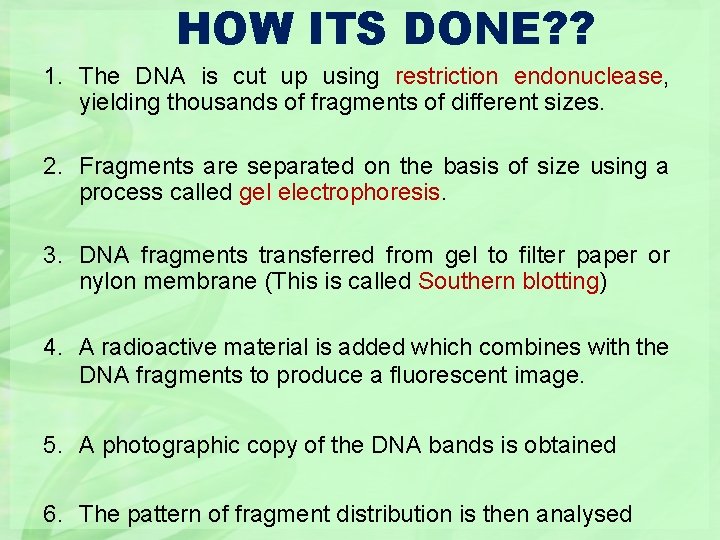
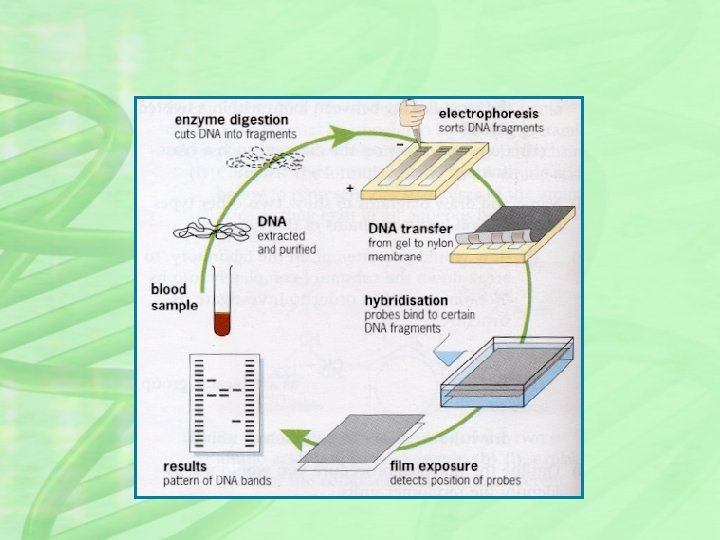
HOW ITS DONE? ? 1. The DNA is cut up using restriction endonuclease, yielding thousands of fragments of different sizes. 2. Fragments are separated on the basis of size using a process called gel electrophoresis. 3. DNA fragments transferred from gel to filter paper or nylon membrane (This is called Southern blotting) 4. A radioactive material is added which combines with the DNA fragments to produce a fluorescent image. 5. A photographic copy of the DNA bands is obtained 6. The pattern of fragment distribution is then analysed


For many years, most DNA profiles contained specific banding patterns on Southern blots of genomic DNA cleaved with a specific restriction enzyme and hybridized to appropriate DNA probes Today, most DNA profiles are electropherograms produced by using PCR primers tagged with fluorescent dyes to amplify the genomic DNA segments of interest, capillary gel electrophoresis to separate the PCR products, and lasers and photocells (fluorescence detectors) to record the sizes of the fluorescent PCR products. The separation and detection steps are performed using the automated DNA sequencing machines.

APPLICATIONS • • • evidence in criminal cases paternity tests immigration requests studying biodiversity tracking genetically modified crops

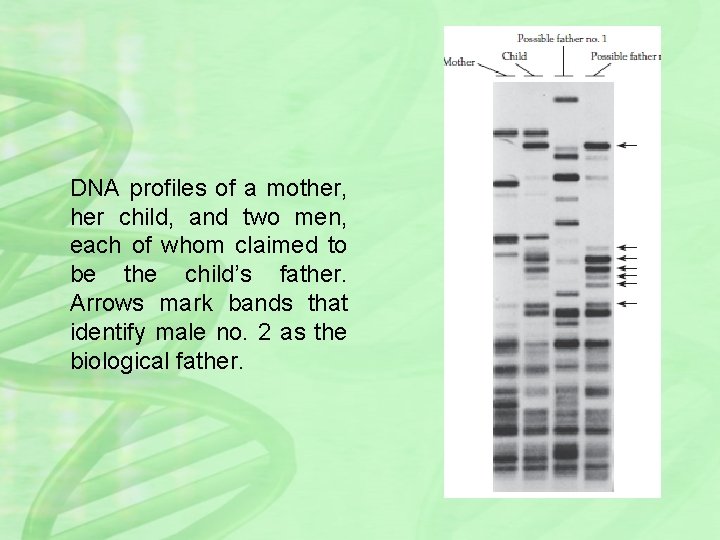
DNA profiles of a mother, her child, and two men, each of whom claimed to be the child’s father. Arrows mark bands that identify male no. 2 as the biological father.

Famous cases • In 2002 Elizabeth Hurley used DNA profiling to prove that Steve Bing was the father of her child Damien

DNA profiles were first used as evidence in a criminal case in 1988. In 1987, a Florida judge denied the prosecutor’s request to present statistical interpretations of DNA evidence against an accused rapist. After a mistrial, the suspect was released. Three months later, he was again in court, accused of another rape. This time the judge allowed the prosecutor to present a statistical analysis of the data based on appropriate population surveys. The analysis showed that the DNA profiles prepared from semen recovered from the victim had a probability of about one in 10 billion of matching the DNA profile of the suspect purely by chance. This time the suspect was convicted. There can be no question about the value of DNA profiles in forensic cases of this type when good tissue or cell samples are obtained from the scene of the crime. If performed carefully by trained scientists and interpreted conservatively using valid population-based data on the frequencies of the polymorphisms involved, DNA profiles can provide a much-needed and powerful tool in the ongoing fight against crime.

Famous cases • Colin Pitchfork was the first criminal caught based on DNA fingerprinting evidence. • He was arrested in 1986 for the rape and murder of two girls and was sentenced in 1988

Famous cases • O. J. Simpson was cleared of a double murder charge in 1994 which relied heavily on DNA evidence. • This case highlighted lab difficulties

Ground zero after the collapse of the Twin Towers of the World Trade Center on September 11, 2001. The bodies of some of the nearly 3000 people killed in the collapse could be identified only by comparing their DNA sequences with those of close relatives, a process called DNA profiling.

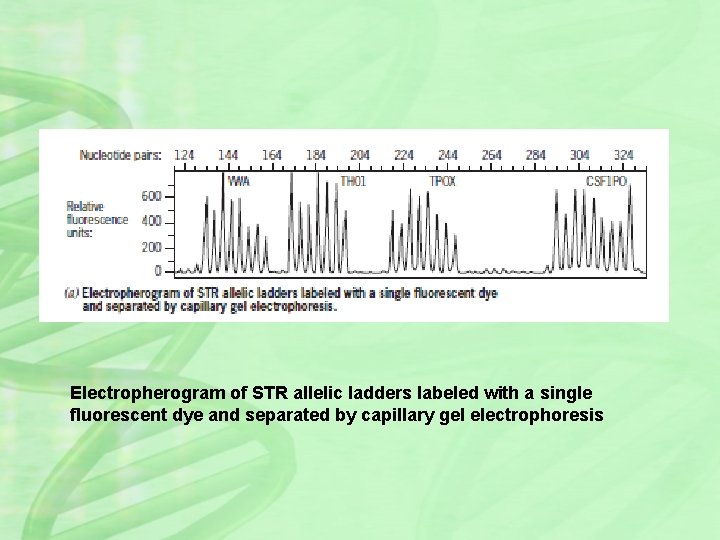
Electropherogram of STR allelic ladders labeled with a single fluorescent dye and separated by capillary gel electrophoresis

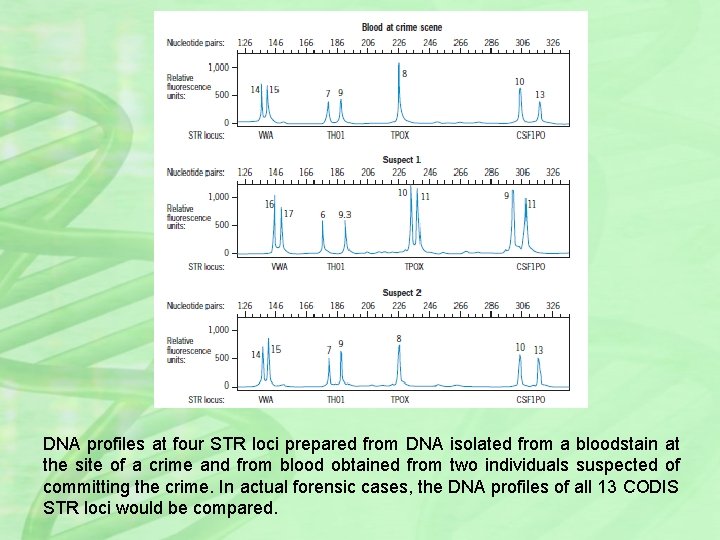
DNA profiles at four STR loci prepared from DNA isolated from a bloodstain at the site of a crime and from blood obtained from two individuals suspected of committing the crime. In actual forensic cases, the DNA profiles of all 13 CODIS STR loci would be compared.

Type of STR profiles used in forensic cases. For the sake of simplicity, the DNA profiles are shown for only 4 of the standard 13 CODIS STR loci. In practice, the profiles of all 13 loci would be compared. The DNA profile prepared from the bloodstain at the crime scene matches the DNA profile from suspect 2, but not the profile from suspect 1. Of course, these matching DNA profiles by themselves do not prove that suspect 1 committed the crime, but, if combined with additional DNA profiles and supporting evidence, they provide strong evidence that suspect 1 was at the scene of the crime. Perhaps more importantly, these profiles clearly show that the blood cells in the stain were not from suspect 1. Thus, DNA profiles have proven invaluable in reducing the frequency of wrongful convictions, and in several cases they have exonerated prisoners in jail for crimes they did not commit. By comparing STR profiles at all 13 CODIS loci, perhaps supplemented with mitochondrial DNA evidence, the possibility that DNA profiles from two individuals will match just by chance can virtually be eliminated. Indeed, the chance that two unrelated Caucasians in a randomly mating population will have identical DNA profiles at all 13 CODIS loci is approximately one in 5. 75 trillion. Clearly, DNA profiling is a powerful tool in personal identity cases.

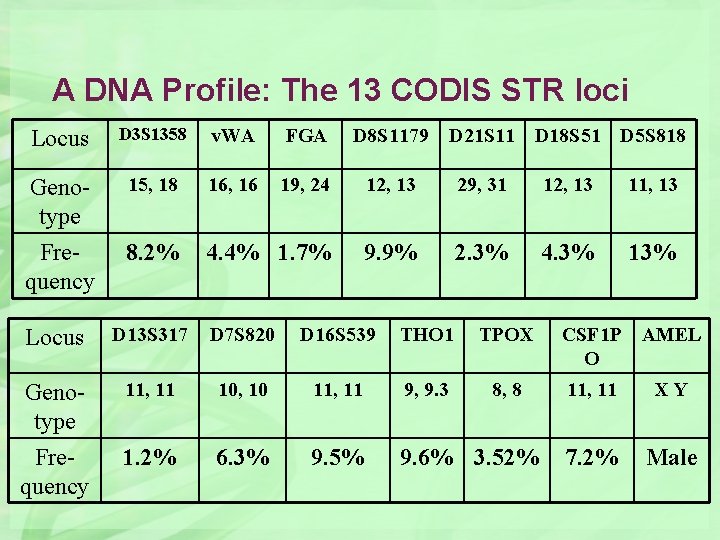
A DNA Profile: The 13 CODIS STR loci Locus D 3 S 1358 v. WA FGA D 8 S 1179 Genotype 15, 18 16, 16 19, 24 12, 13 29, 31 12, 13 11, 13 Frequency 8. 2% 4. 4% 1. 7% 9. 9% 2. 3% 4. 3% 13% Locus D 13 S 317 D 7 S 820 D 16 S 539 THO 1 TPOX CSF 1 P O AMEL Genotype Frequency 11, 11 10, 10 11, 11 9, 9. 3 8, 8 11, 11 XY 1. 2% 6. 3% 9. 5% 9. 6% 3. 52% 7. 2% Male D 21 S 11 D 18 S 51 D 5 S 818

How common or rare would this 13 locus DNA profile be in the reference population? In most cases, a "product rule" calculation can be done by multiplying each individual probability together By combining the frequency information for all 13 CODIS loci, this frequency of this profile would be 1 in 7. 7 quadrillion Caucasians…that’s 1 in 7. 7 x 1015 power!