DNA elements distinguished by chromatin structure ENCODE datatypes
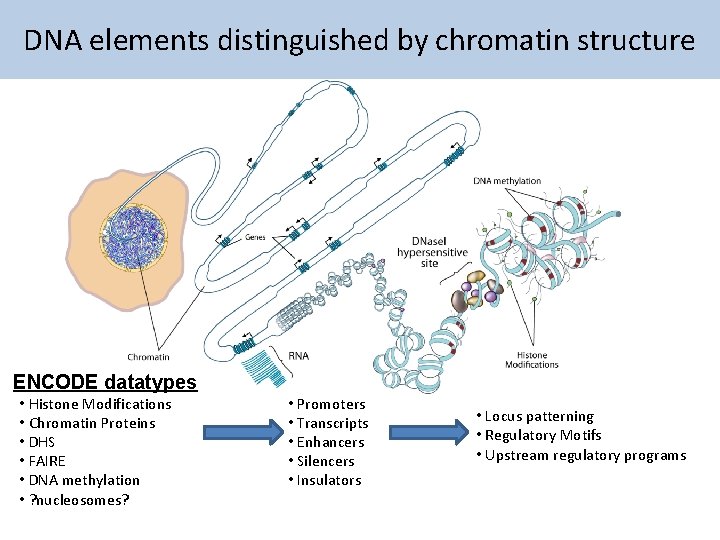
DNA elements distinguished by chromatin structure ENCODE datatypes • Histone Modifications • Chromatin Proteins • DHS • FAIRE • DNA methylation • ? nucleosomes? • Promoters • Transcripts • Enhancers • Silencers • Insulators • Locus patterning • Regulatory Motifs • Upstream regulatory programs

Integration Vignettes for Chromatin 1. Functional elements: A. What ENCODE features are correlated with specific structural features of gene (TSS, TTS, etc. )? B. Can different biochemical events be used to predict (discover) chromosomal features? C. Enhancers show similar chromatin structure to promoters D. What is the nature of regions that are devoid of signal by ENCODE methods, the "dead zones"?
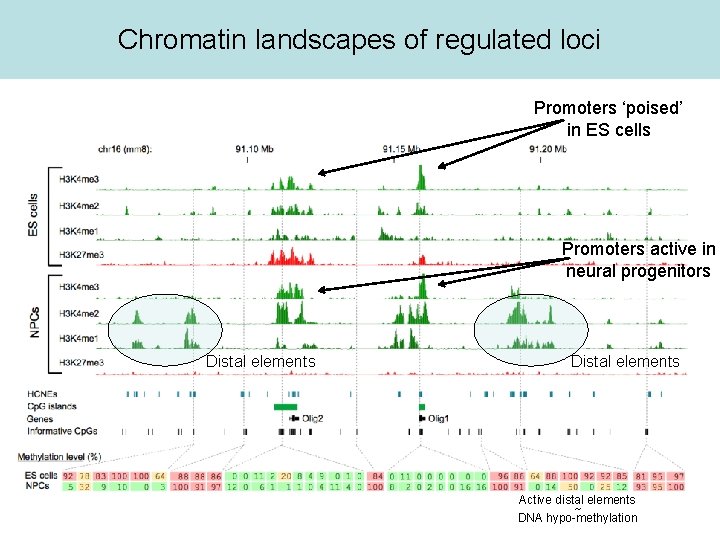
Chromatin landscapes of regulated loci Promoters ‘poised’ in ES cells Promoters active in neural progenitors Distal elements Active distal elements ~ DNA hypo-methylation
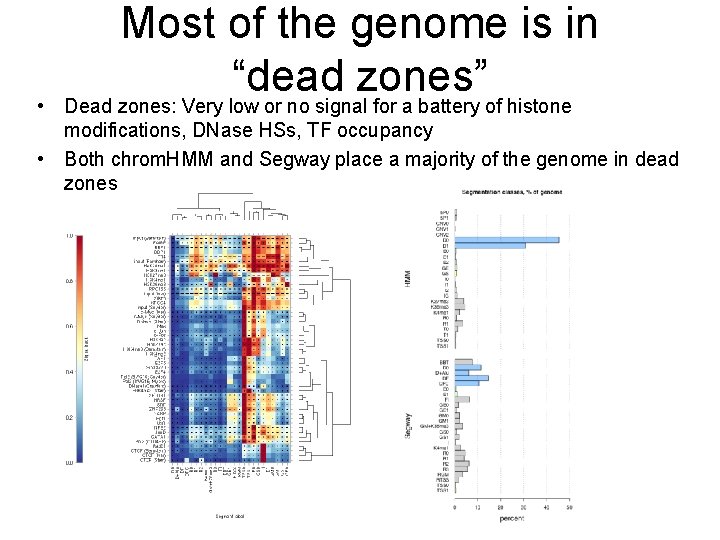
Most of the genome is in “dead zones” • Dead zones: Very low or no signal for a battery of histone modifications, DNase HSs, TF occupancy • Both chrom. HMM and Segway place a majority of the genome in dead zones
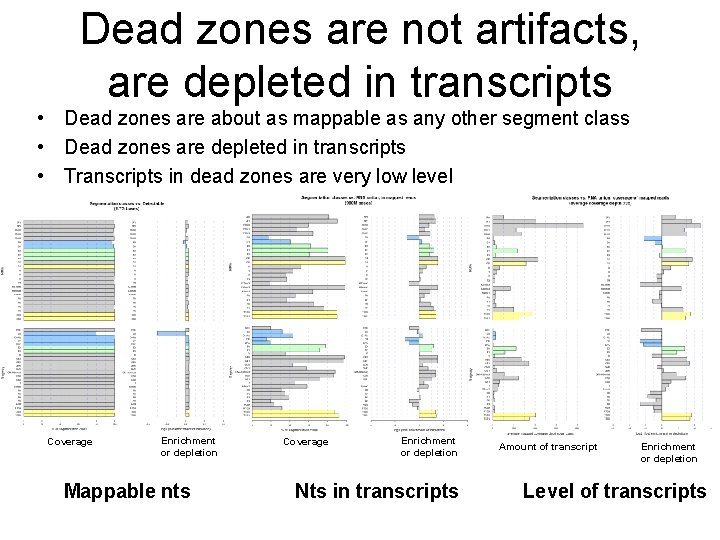
Dead zones are not artifacts, are depleted in transcripts • Dead zones are about as mappable as any other segment class • Dead zones are depleted in transcripts • Transcripts in dead zones are very low level Coverage Enrichment or depletion Mappable nts Coverage Enrichment or depletion Nts in transcripts Amount of transcript Enrichment or depletion Level of transcripts

Why are dead zones important? • Interpretation – Dead zones are condensed chromatin (heterochromatin) that enzymes have little or no access to – A substantial part of the genome, including inactive genes, are in this chromatin state • Implication: – One major mechanism for turning genes off and keeping them off is to put them into this highly condensed state – This is distinct from turning genes off via Polycomb (PRC 2, H 3 K 27 me 3), H 3 K 9 me 2, 3 and HP 1, etc.
Integration Vignettes for Chromatin 2. Other ENCODE assays or DNA sequence features that correspond to chromatin landscape A. Are ENCODE assays correlated in specific systematic patterns across the genome? Tx, TFs B. The exceptions? Punctate histone marks (k 4 me 1) ≠ Dnase/FAIRE; TFs ≠ to open chromatin C. Accessibility, nucleosome positions D. Different classes of repetitive elements

Integration Vignettes for Chromatin 3. TF binding events (what can chromatin landscape teach us? ) A. Biochemical features associated with different TF binding events? Are there classes of TFs? B. Are there different patterns of DNase. I footprinting between factors C. How do TF binding events affect or are guided by nucleosome positioning?
Integration Vignettes for Chromatin 4. Higher-level biological questions? A. TF binding events that correlate with open chromatin BECAUSE they trigger nucleosome remodeling. B. TF binding events that correlate with open chromatin BECAUSE they can only bind accessible DNA. C. How predictive are ENCODE assays at predicting differential gene expression in different cell types?
- Slides: 9