DNA damage response and chromatin remodeling Presentation for









- Slides: 36

DNA damage response and chromatin remodeling Presentation for Ron Shamir group internal meeting 3/11/2010

DNA damages, including single- and double- strand breaks (SSB and DSB respectively), pyrimidine dimmers (PD), and oxidized nucleotides, are caused by physical agents in the environment, like ionizing radiation (IR) and ultra violet light (UV), and by various chemicals (including reactive oxygen species (ROS) from internal metabolic processes source).

Ø In eukaryotes, the sensing and repair of DNA damages occur in chromatin context. Chromatin constitutes a physical barrier to enzymes and regulatory factors to reach the DNA. To deal with this obstacle, transient chromatin structural changes are integral to base- and nucleotide-excision, homologous recombination, and non-homologous end joining DNA repair pathways.

DDR and Chromatin Remodeling


Nucleotide Excision Repair (NER)

Ø Base Excision Repair (BER)

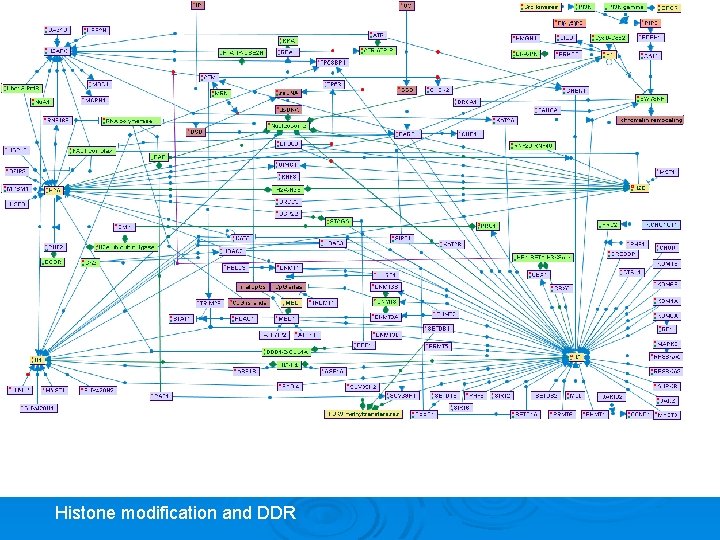
HDAC (Histone Deacetylase) Families Class I and II: • Zinc-hydrolase • ~11 human homologs • Inhibited by hydroxamic acid containing compounds (TSA, SAHA) Class III (Sir 2 or sirtuins) • No structural/mechanistic similarity to Class I and II • NAD+ requiring • 7 human homologs • Inhibited by nicotinamide Histone modification and DDR

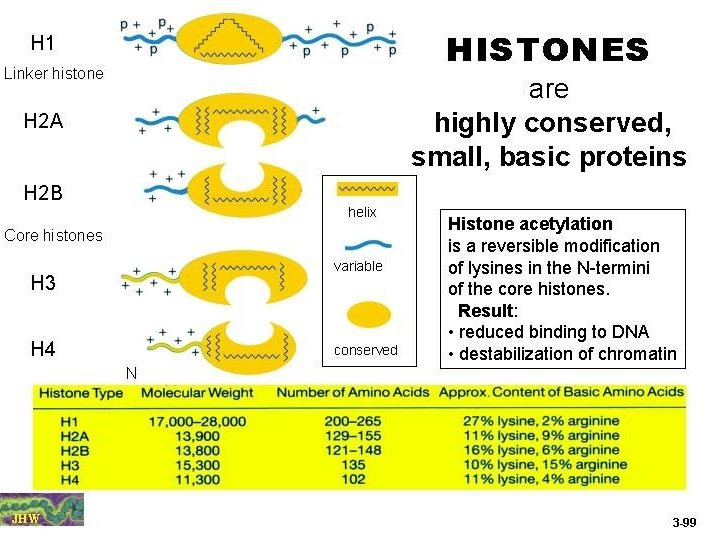
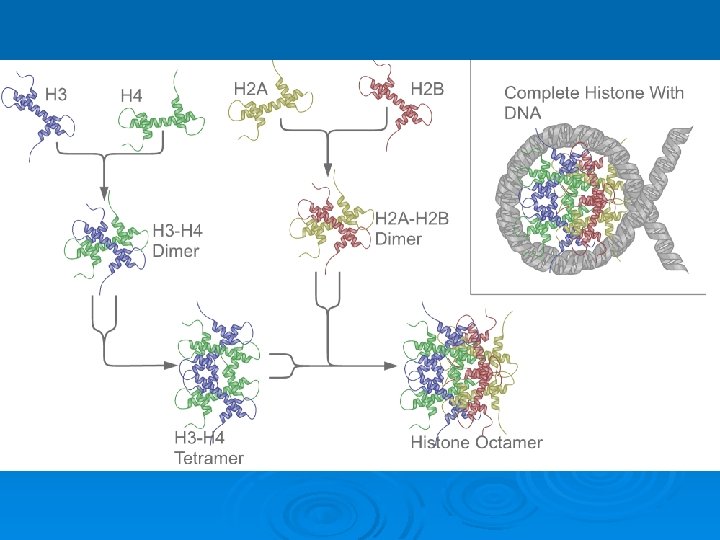
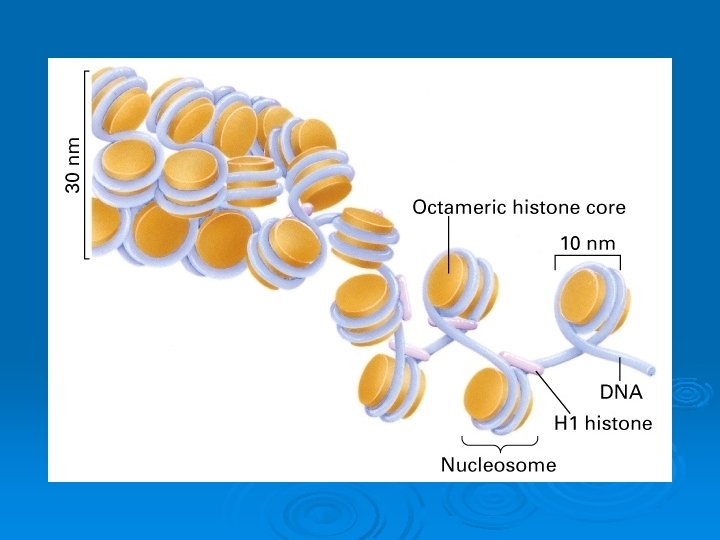
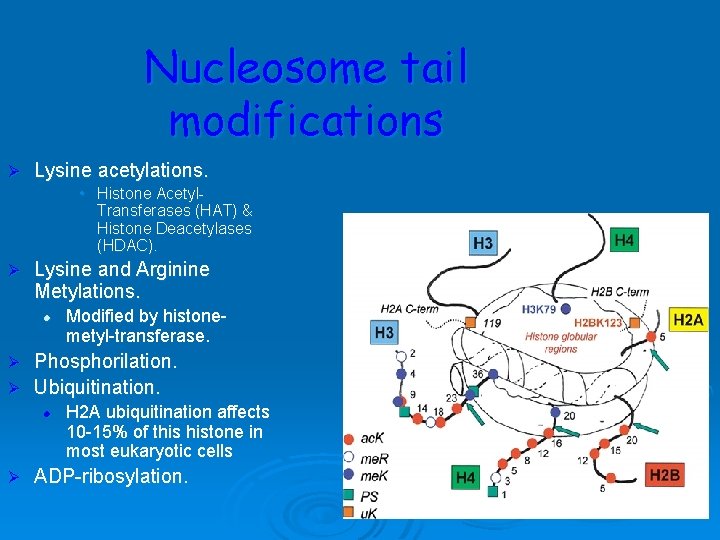
The basic structural subunits of the chromatin are the nucleosomes. These core particles consists of about 146 bp of ds. DNA wrapped in 1. 65 left-handed superhelical turns around four identical pairs of the histone proteins H 2 A, H 2 B, H 3 and H 4, collectively known as the histone octamer. The remaining 50 bp of the repeating unit consists of "linker DNA", ds. DNA which separates the core particles.


Histone-histone associations The four core histones tend to associate with each other in very specific patterns of interactions. Histones H 3 and H 4 interact very strongly with each other and form a specific complex, a tetramer. Histones H 2 A and H 2 B also interact with each other and can form primarily dimers and some higher oligomers. The pattern of association is: H 3 H 4 H 2 A H 2 B


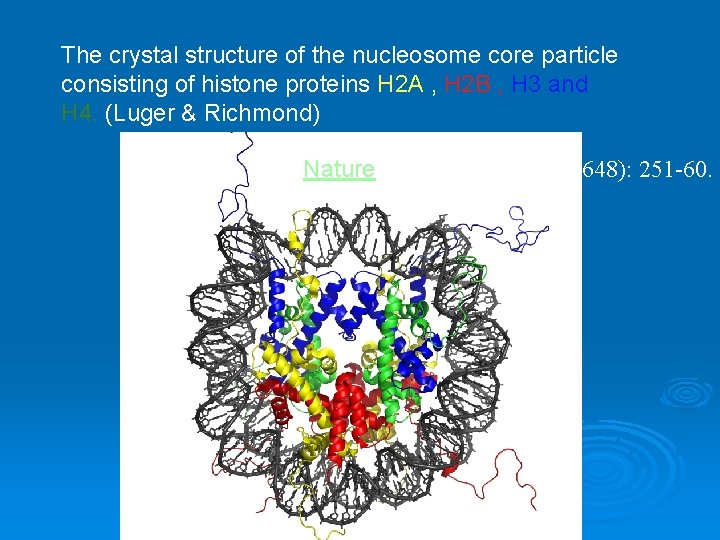
The crystal structure of the nucleosome core particle consisting of histone proteins H 2 A , H 2 B , H 3 and H 4. (Luger & Richmond) Nature. 1997 Sep 18; 389 (6648): 251 -60.

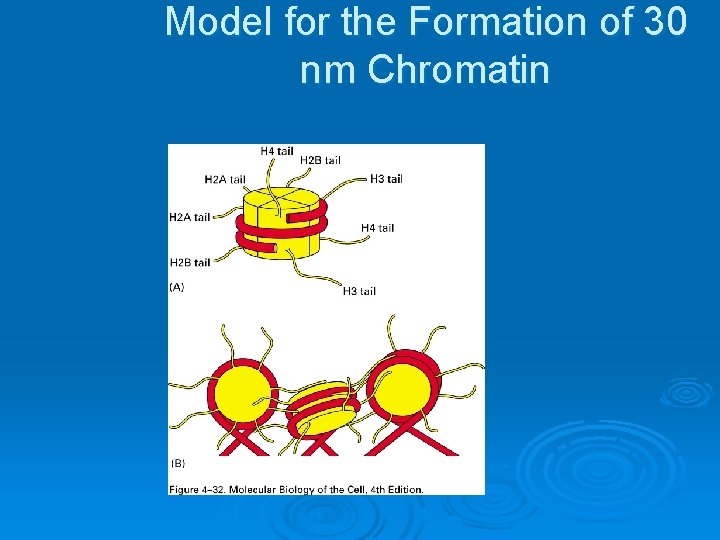
Model for the Formation of 30 nm Chromatin

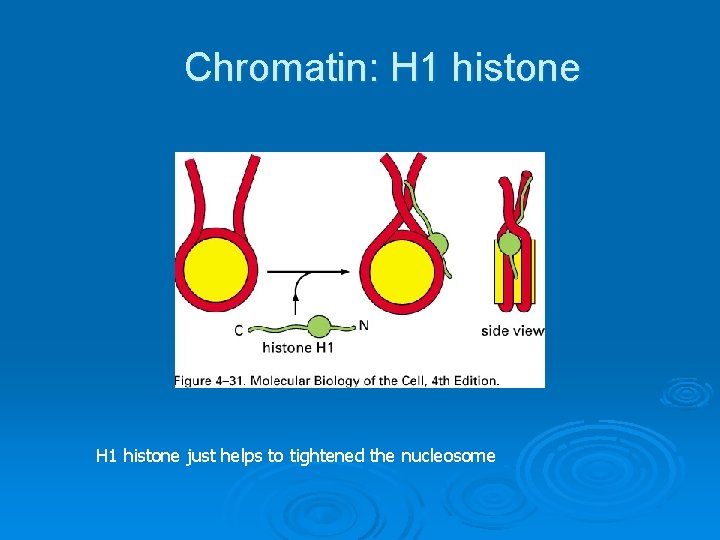
Chromatin: H 1 histone just helps to tightened the nucleosome


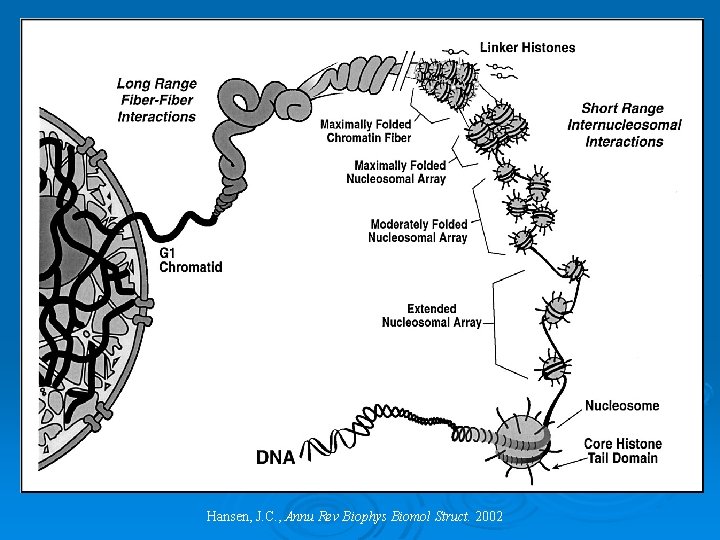
The Organization of Chromatin Hansen, J. C. , Annu Rev Biophys Biomol Struct. 2002

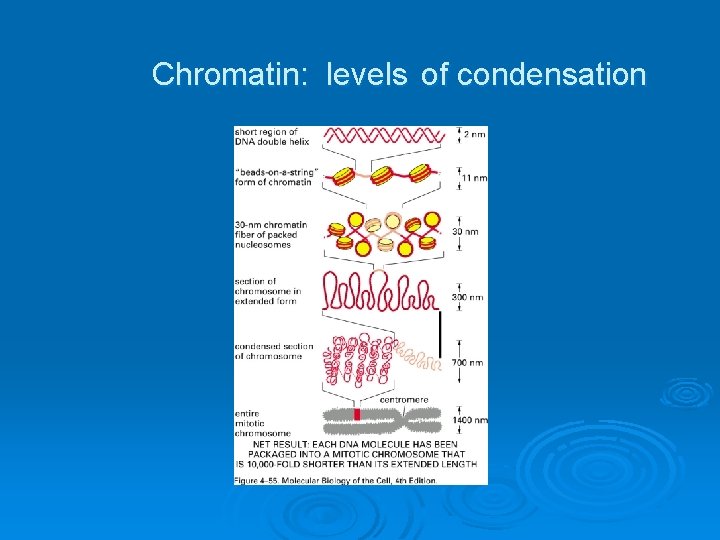
Chromatin: levels of condensation

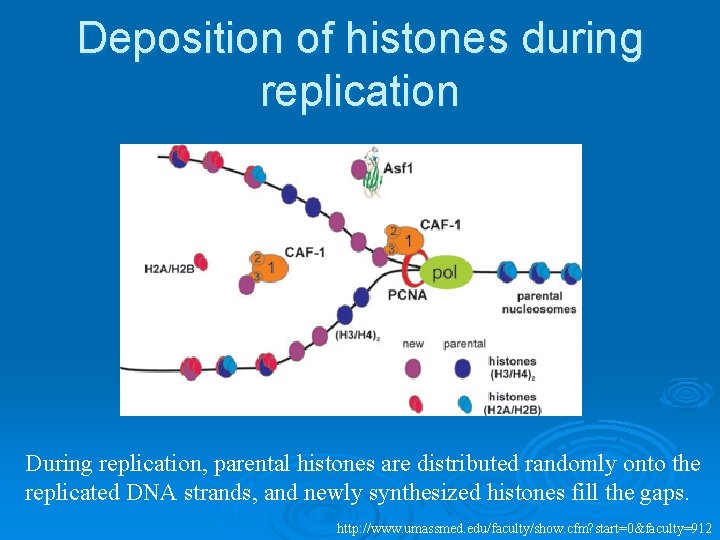
Deposition of histones during replication During replication, parental histones are distributed randomly onto the replicated DNA strands, and newly synthesized histones fill the gaps. http: //www. umassmed. edu/faculty/show. cfm? start=0&faculty=912

Ø Chromatin remodeling mechanisms includes: Ø (1) posttranslational modifications (PTMs) of histones and non-histones chromatin associated proteins, including covalent additions (removals) of phosphoryl-, methyl-, acetyl-, ubiquitin-, SUMO, and ADP-ribose Ø (2) histone-variants replacement, and nucleosomes repositioning or eviction. ATP-dependent multi subunits chromatin remodeling complexes (INO 80, SRCAP/SWRC, SWI/SNF, and MI-2/Nu. RD) implement the latter mechanisms

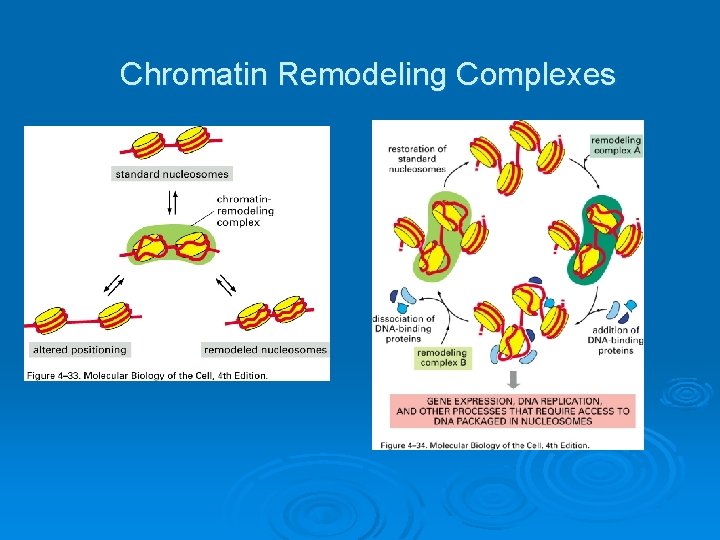
Enzymes that regulate chromatin Two classes of enzymes that regulate chromatin structure: histone modifiers and chromatin remodelers. Ø Histone modifiers don’t alter nucleosome position; they make passive marks that recruit more active functions (histone code). Ø Chromatin remodelers hydrolyze ATP to actively remodel chromatin: shift nucleosome position with respect to DNA, exposing or occluding regulatory sequences. Ø These enzymes function within larger complexes of subunits that collectively act to enhance and/or target the remodeling activity. Ø

ATP-dependant chromatin remodeling complexes Ø SWI/SNF: 15 subunits complex. Catalytic subunits are SMARCA 2 or SMARCA 4. plays essential roles in a variety of cellular processes including differentiation, proliferation and DNA repair. Ø SRCAP/SWR-C: 10 subunits complex. incorporate the histone variant H 2 AFZ into nucleosomes. Ø INO 80: 16 -subunits complex. Involved in DNA repair, checkpoint regulation, and DNA replication, cooperatively with their histone substrates, gamma-H 2 AX and H 2 AFZ. Ø Mi-2/Nu. RD: 13 subunits complex. The Catalytic subunits are CHD 3 and CHD 4. Involved in the regulation of some important DDR genes like P 53, BRCA 1 and MCPH 1.

Histone PTMs influence genome function by: Ø directly disturbing nucleosome stability which affects chromatin compaction and accessibility Ø constituting a docking site for different kinds of non-histone proteins Ø Certain histone PTMs indicate the specific position of DNA damage and provide a platform of interaction for DDR proteins, determining the repair pathway that should be involved

Ø Ø The best-known histone PTM is histone acetylation deacetylation, controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Histone acetylations enhance chromatin accessibility. and facilitate DNA repair beyond their well-documented role in transcription, presumably by opening or loosening compact nucleosomal structure close to sites of damage. Histone phosphorylation mediated by members of the PI 3 K kinase superfamily (ATM, ATR, DNA-PK) plays a role at the beginning of DDR by facilitating the access of different repair proteins to DNA breaks

Nucleosome tail modifications Ø Lysine acetylations. • Histone Acetyl. Transferases (HAT) & Histone Deacetylases (HDAC). Ø Lysine and Arginine Metylations. l Modified by histonemetyl-transferase. Phosphorilation. Ø Ubiquitination. Ø l Ø H 2 A ubiquitination affects 10 -15% of this histone in most eukaryotic cells ADP-ribosylation.

Acetylation of Lysine 16 of histone H 4 completely abolishes the ability of the tail domains to mediate nucleosome interactions, which are required for chromatin condensation.

Chromatin remodeling is an active process Chromatin remodeling describes the energydependent (ATP) displacement or reorganization of nucleosomes that occurs in conjunction with activation (or repression) of genes for transcription Ø Chromatin remodelers also play a role in recombination and repair Ø

Chromatin Remodeling Complexes

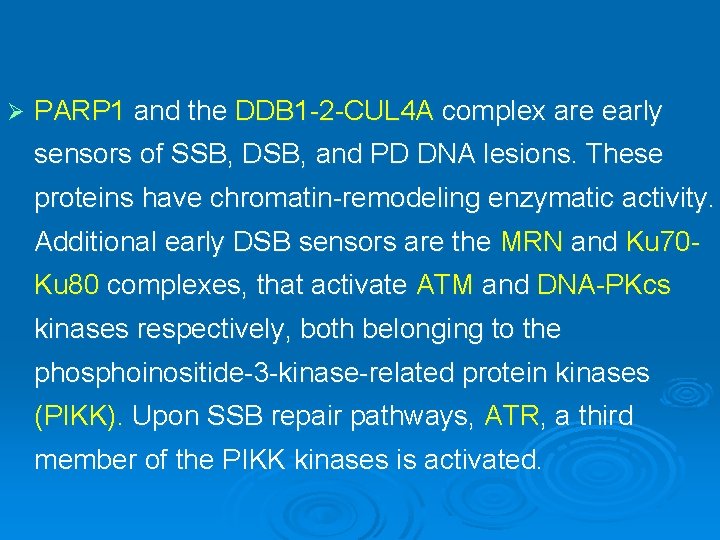
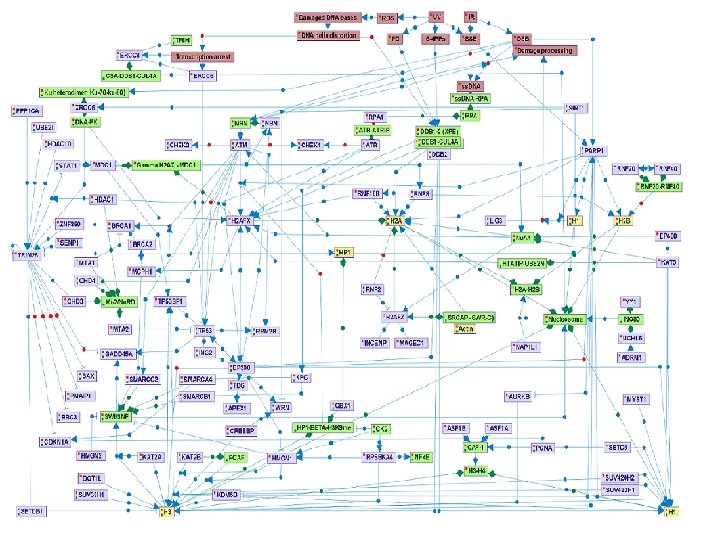
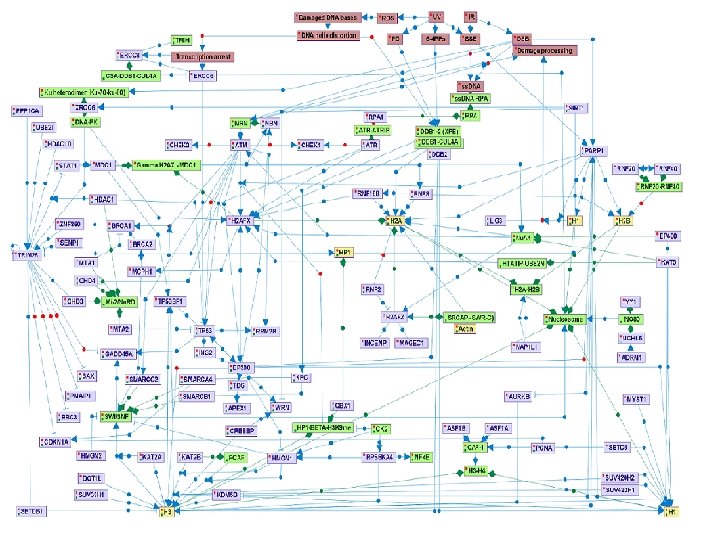
Ø PARP 1 and the DDB 1 -2 -CUL 4 A complex are early sensors of SSB, DSB, and PD DNA lesions. These proteins have chromatin-remodeling enzymatic activity. Additional early DSB sensors are the MRN and Ku 70 Ku 80 complexes, that activate ATM and DNA-PKcs kinases respectively, both belonging to the phosphoinositide-3 -kinase-related protein kinases (PIKK). Upon SSB repair pathways, ATR, a third member of the PIKK kinases is activated.


Ø The PIKK kinases phosphorylate many substrates, including histones and proteins involved in regulation of chromatin structure. One of these proteins is TRIM 28 (KAP 1), a transcription co-repressor proposed to regulate chromatin structure and heterochromatin formation. In response to the induction of DNA DSBs, its co-repression function is inhibited by ATMdependant phospohorylation.

Ø The main histone PTMs in response to DSBs are those related to H 2 AFX (histone H 2 A variant). DDR-related replacement of H 2 A variant in the nucleosomes by H 2 AFZ (promoted by the SRCAP/SWR-C ATPdependent complex) also occur.


Ø Upon DSB, the histone acetylase complex NUA 4/TIP 60, several histones ubiquitin ligases are activated. The interactions between the DDR proteins, MCPH 1, P 53 and BRCA 1, and components of the SWI/SNF and MI-2/Nu. RD chromatin remodeling complexes, seems to be important to the regulation of DDR.

In conclusion: In eukaryotic cells, packaging of DNA into highly condensed chromatin presents a significant obstacle to DNA-based processes. (replication, repair and recombination). Cells use three major strategies to allows protein factors to gain access to nucleosomal DNA: 1. Histone posttranslational modifications (PTMs) 2. Incorporation of histone variants into nucleosomes 3. ATP-dependent chromatin remodeling

Ø ATP-dependent multi subunits chromatin remodeling complexes (INO 80, SRCAP/SWRC, SWI/SNF, and MI-2/Nu. RD) implement the latter mechanisms and many proteins that are involved in DNA repair and DDR have physical interactions and reciprocal regulations with some proteins that belongs these complexes.