DNA Computing on a Chip Mitsunori Ogihara and






- Slides: 9

DNA Computing on a Chip Mitsunori Ogihara and Animesh Ray Nature, vol. 403, pp. 143 -144 Cho, Dong-Yeon

Abstract l In a DNA computer ¨ The input and output are both strands of DNA. ¨ A computer in which the strands are attached to the surface of a chip can now solve difficult problems quite quickly. l [Liu et al. , 2000] ¨ Liu, Q. et al. , “DNA computing on a chip, ” Nature, vol. 403, pp. 175 -179, 2000.

Arriving at the truth by elimination l Problem classes ¨ Polynomial time or P problems < O(1), O(n), O(nlogn), O(n 2), O(n 3), … ¨ Non-deterministic polynomial time or NP problems < ‘Hard’ NP problems have running times that grow exponentially with the number of the variables. < O(2 n), O(3 n), O(n!) … l New technology for massively parallel elimination [Liu et al. , 2000] ¨ This algorithm harnesses the power of DNA chemistry and biotechnology to solve a particularly difficult problem in mathematical logic.

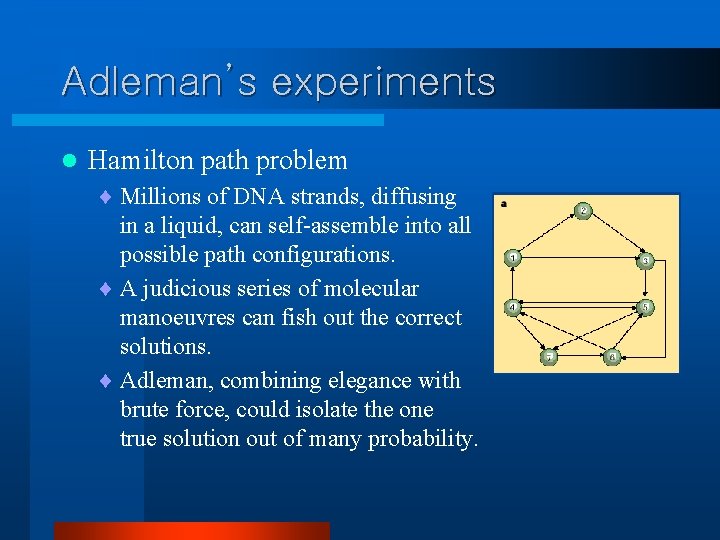
Adleman’s experiments l Hamilton path problem ¨ Millions of DNA strands, diffusing in a liquid, can self-assemble into all possible path configurations. ¨ A judicious series of molecular manoeuvres can fish out the correct solutions. ¨ Adleman, combining elegance with brute force, could isolate the one true solution out of many probability.

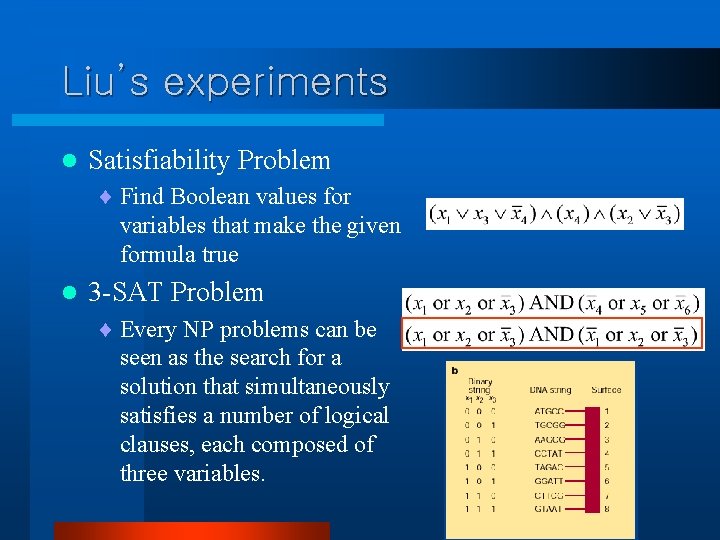
Liu’s experiments l Satisfiability Problem ¨ Find Boolean values for variables that make the given formula true l 3 -SAT Problem ¨ Every NP problems can be seen as the search for a solution that simultaneously satisfies a number of logical clauses, each composed of three variables.

Procedure l Step 1. ¨ Attach DNA strings encoding all possible answers to a specially treated gold surface. l Step 2. ¨ Complementary DNA strands that satisfy the first clauses are added to the solution. ¨ The remaining single strands are destroyed by enzymes. ¨ The surface is then heated to melt away the complementary strands. ¨ This cycle is repeated for each of the remaining clauses.

l Step 3. ¨ The surviving strands first have to be amplified using the PCR. ¨ Their identities are then determined by pairing with an ordered array of strings identical to the original set of sequences. l O(3 k+1) vs. O(1. 33 n), O(2 n) ¨ k: the number of clauses ¨ n: the number of variables

Problems l Scaling up this technique to solve larger 3 -SAT problems is still unrealistic. ¨ Correcting errors arising from the inherent sloppiness of DNA chemistry ¨ High cost of tailor-made DNA sequences < 50 -variable 3 -SAT: 1015 unique DNA strands ¨ Designing enough unique DNA strands ¨ Exponentially increasing number of DNA molecules <A compromise may be achieved by reducing the search space through heuristics.

Conclusions l The ideal application for DNA computation does not lie in computing large NP problems ¨ There may be a need for fully organic computing devices implanted within a living body that can integrated signals from several sources and compute a response in terms of an organic molecular-delivery device for a drug or signal.