DNA Classifications with SelfOrganizing Maps SOMs Thanakorn Naenna





- Slides: 24

DNA Classifications with Self-Organizing Maps (SOMs) Thanakorn Naenna Mark J. Embrechts Robert A. Bress May 2003 IEEE International Workshop on Soft Computing in Industrial Application 1

Presentation Outline • • • Introduction to DNA Splice Junctions Data Collection Introduction to SOMs SOM for DNA Splice Junction Classification Results Conclusions 2

3

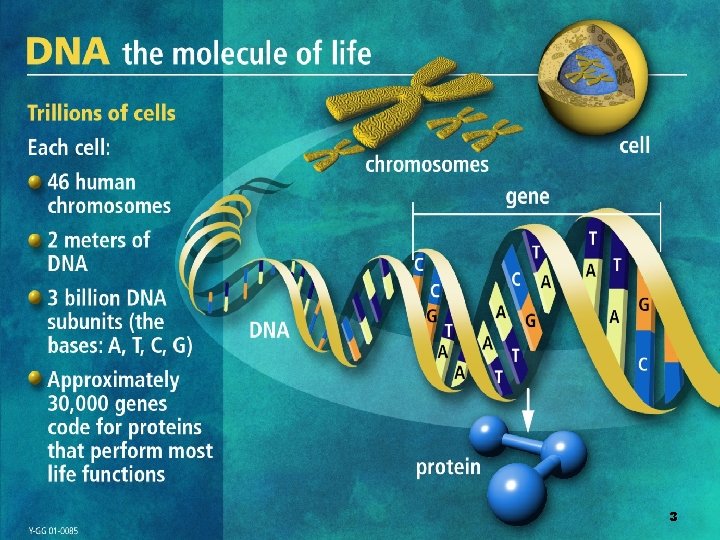
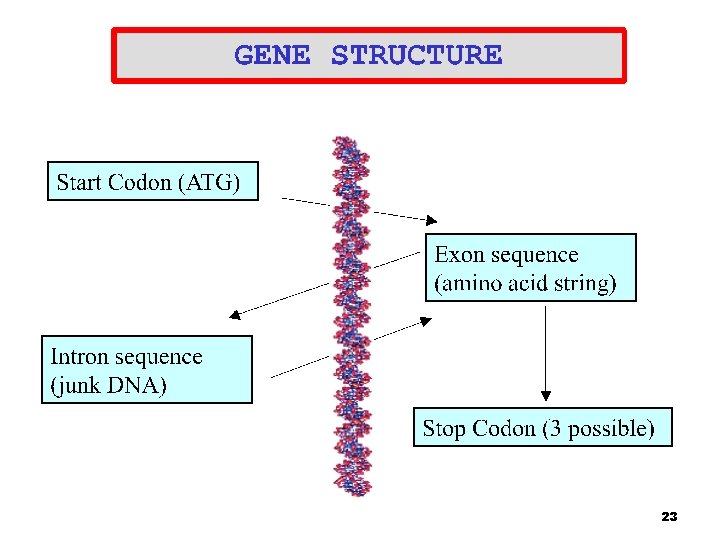
Human genome in a nutshell • • • Human : 23 chromosomes Chromosomes thousands of genes Gene info : exons , comments : introns Splice junction are like /* comment flags */ in C-code Exons and introns codons Codon bases 4

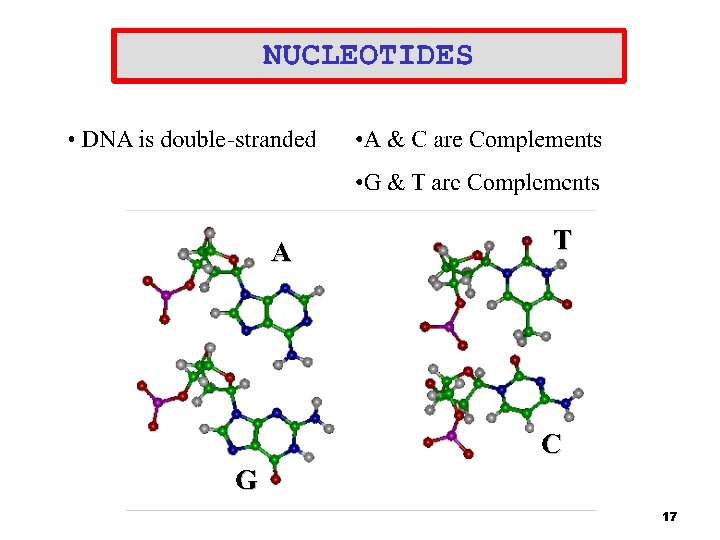
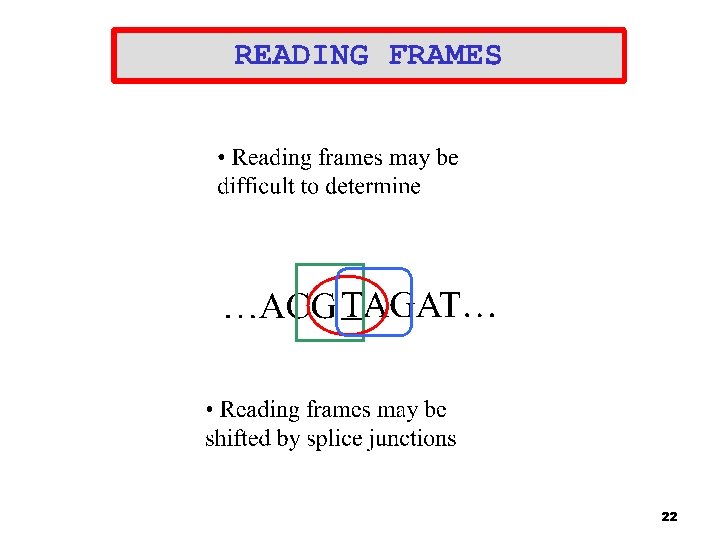
DNA Splice Junctions • • DNA billions of nucleotides ( A, C, G, T) Genes sequences of amino acids (exons) that are often interrupted by non-coding nucleotides (introns) <. 1% of human DNA is made up of exons 99% of splice junctions have the same motif, for Exon to intron it is GT – Intron to exon it is AG – Intron Splice Junction Exon Splice Junction Intron …. GTGAAGGTTAA AGATGTAGAT GT ATTG… 5

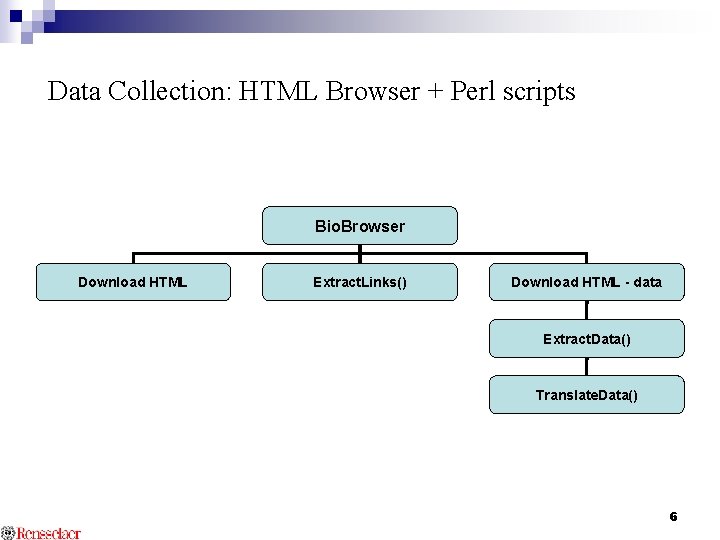
Data Collection: HTML Browser + Perl scripts Bio. Browser Download HTML Extract. Links() Download HTML - data Extract. Data() Translate. Data() 6

7

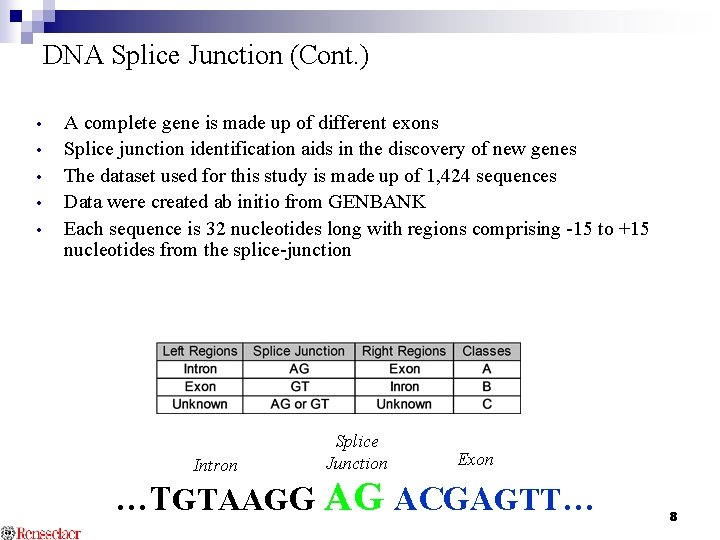
DNA Splice Junction (Cont. ) • • • A complete gene is made up of different exons Splice junction identification aids in the discovery of new genes The dataset used for this study is made up of 1, 424 sequences Data were created ab initio from GENBANK Each sequence is 32 nucleotides long with regions comprising -15 to +15 nucleotides from the splice-junction Intron Splice Junction Exon …TGTAAGG AG ACGAGTT… 8

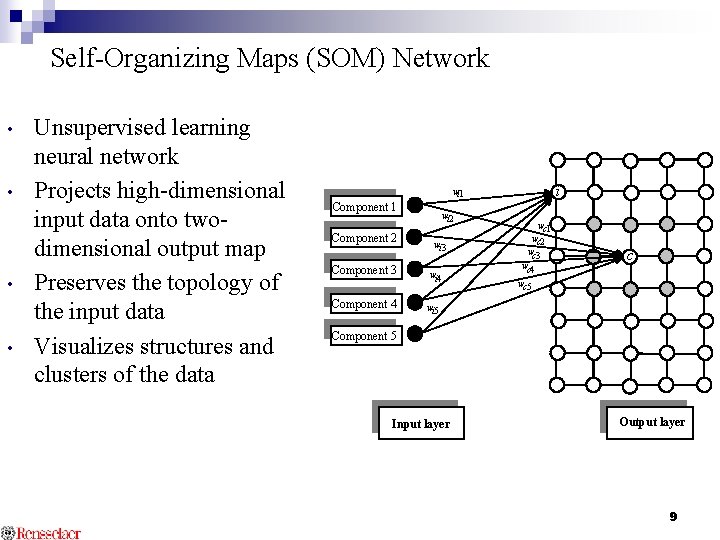
Self-Organizing Maps (SOM) Network • • Unsupervised learning neural network Projects high-dimensional input data onto twodimensional output map Preserves the topology of the input data Visualizes structures and clusters of the data i wi 1 Component 2 Component 3 Component 4 wi 2 wi 3 wi 4 wc 1 wc 2 wc 3 wc 4 c wc 5 wi 5 Component 5 Input layer Output layer 9

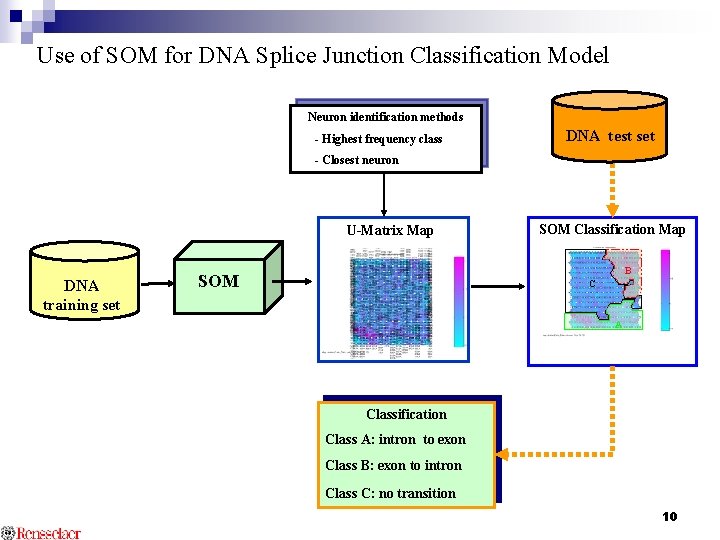
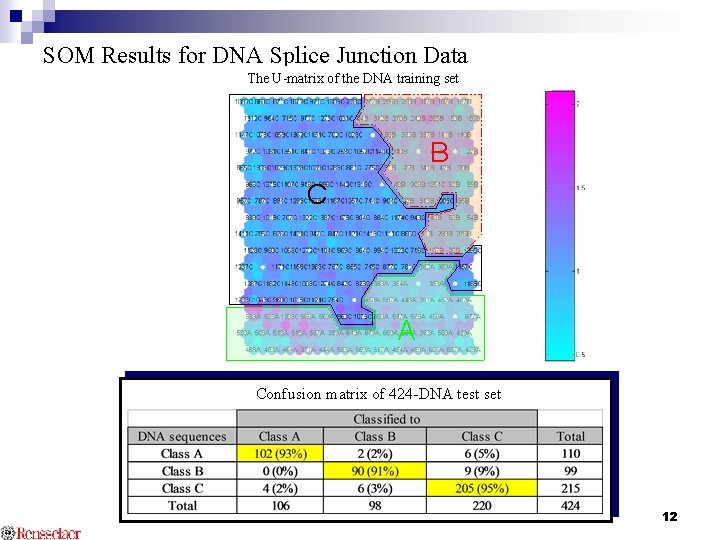
Use of SOM for DNA Splice Junction Classification Model Neuron identification methods - Highest frequency class DNA test set - Closest neuron U-Matrix Map DNA training set SOM Classification Map B SOM C A Classification Class A: intron to exon Class B: exon to intron Class C: no transition 10

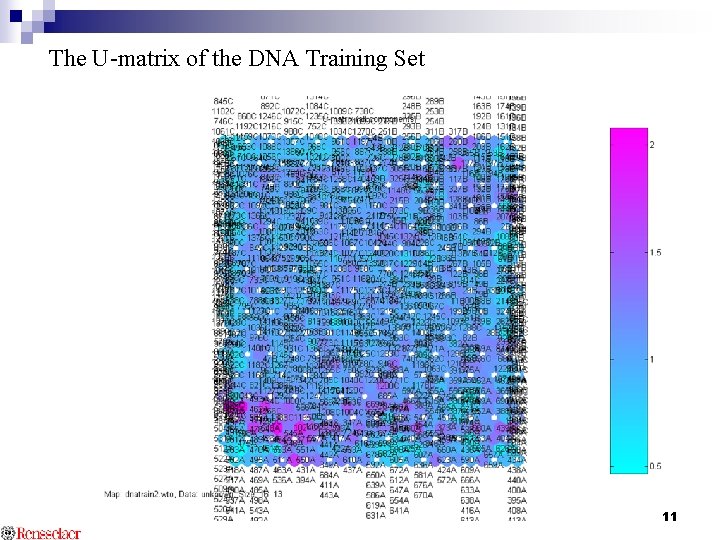
The U-matrix of the DNA Training Set 11

SOM Results for DNA Splice Junction Data The U-matrix of the DNA training set B C A Confusion matrix of 424 -DNA test set 12

Conclusions • • SOM is effective in DNA splice junction classification SOM is powerful visualization for high dimensional data 13

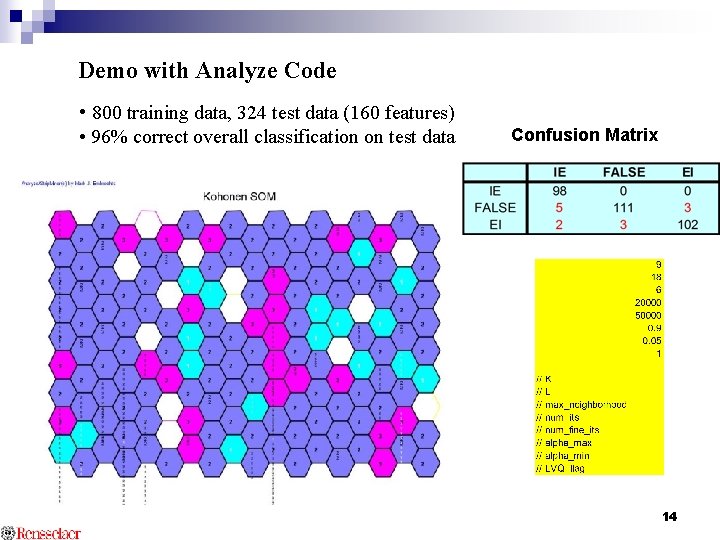
Demo with Analyze Code • 800 training data, 324 test data (160 features) • 96% correct overall classification on test data Confusion Matrix 14

THE END GATCAATGAGGTGGACACCAGAGGCGGGGACTTGTAAATAACACTGGGCTGTAGGAGTGA TGGGGTTCACCTCTAATTCTAAGATGGCTAGATAATGCATCTTTCAGGGTTGTGCTTCTAGAAGGTAGAGCTGTGGTCGTTCAATAAAAGTCCTCAAGAGGTTAATACGCAT GTTTAATAGTACAGTATGGTGACTATAGTCAACAATAATTTATTGTACATTTTTAAATAG CTAGAAGAAAAGCATTGGGAAGTTTCCAACATGAAGAAAAGATAAATGGTCAAGGGAATG GATATCCTAATTACCCTGATTTGATCATTATGCATTATATACATGAATCAAAATATCACA CATACCTTCAAACTATGTACAAATATTATATACCAATAAAAAATCATCATCTCC ATCATCACCACCCTCCTCCTCATCACCACCAGCATCACCACCATCATCACCACCACCATC ATCACCACCACCACTGCCATCATCATCACCACCACTGTGCCATCATCATCACCACCACTG TCATTATCACCACCACCATCATCACCAACACCACTGCCATCGTCATCACCACCACTGTCA TTATCACCACCACCATCACCAACATCACCACCACCATTATCACCACCATCAACACCACCA CCCCCATCATCACTACTACCATCATTACCAGCACCACTATCACCACCACAATCACCACTATCATCAACATCATCACTACCACCATCACCAACACCA CCATCATTATCACCACCACCACCATCACCAACATCACCACCATCATCATCACCACCATCA CCAAGACCATCATCATCACCACCAACATCACCACCATCACCAACACCACCATCA CCACCACCATCATCACCACCATCATCATCACCACCACCGCCATCATCA TCGCCACCACCATGACCACCACCATCACAACCATCACCACCATCACAACCACCATCATCA CTATCGCTATCACCACCATTACCACCACCATTACTACAACCATGACCATCACCACCACCATCACAACGATCACCATCACAGCCACCATCATCACCACCATCAAACCATCGGCATTATTATTTTTTTAGAATTTTGTTGGGATTCAGT ATCTGCCAAGATACCCATTCTTAAAACATGAAAAAGCAGCTGACCCTCCTGTGGCCCCCT TTTTGGGCAGTCATTGCAGGACCTCATCCCCAAGCAGCAGCTCTGGTGGCATACAGGCAA CCCACCACCAAGGTAGAGGGTAATTGAGCAGAAAAGCCACTTCCTCCAGCAGTTCCCTGT 15

16

17

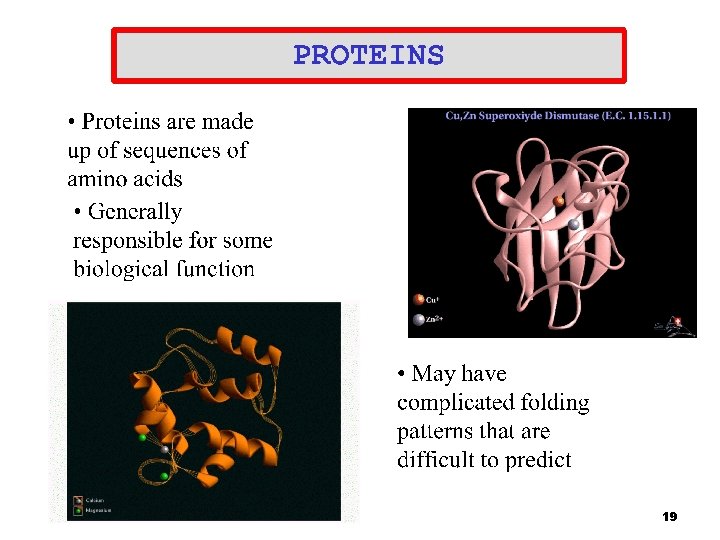
18

19

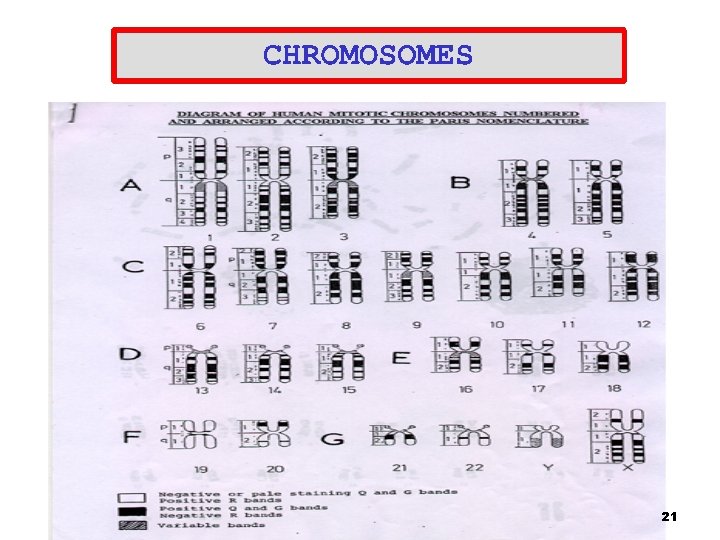
20

21

22

23

24