DNA and RNA Structure Components Sugar Base Phosphate
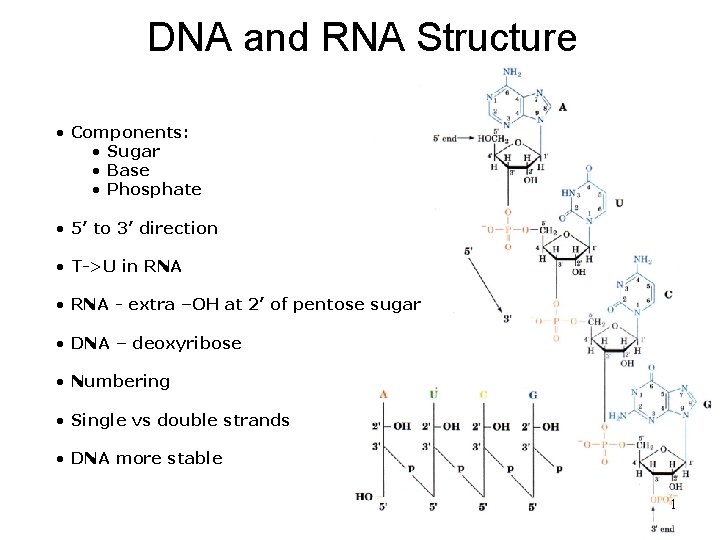
DNA and RNA Structure • Components: • Sugar • Base • Phosphate • 5’ to 3’ direction • T->U in RNA • RNA - extra –OH at 2’ of pentose sugar • DNA – deoxyribose • Numbering • Single vs double strands • DNA more stable 1
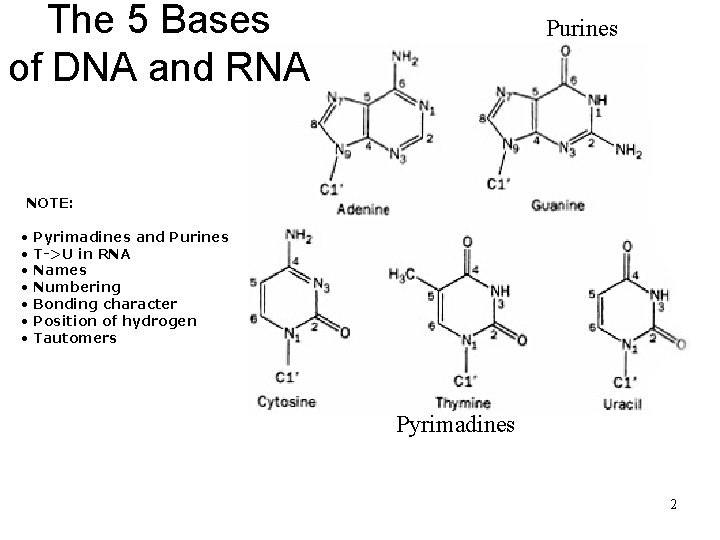
The 5 Bases of DNA and RNA Purines NOTE: • • Pyrimadines and Purines T->U in RNA Names Numbering Bonding character Position of hydrogen Tautomers Pyrimadines 2
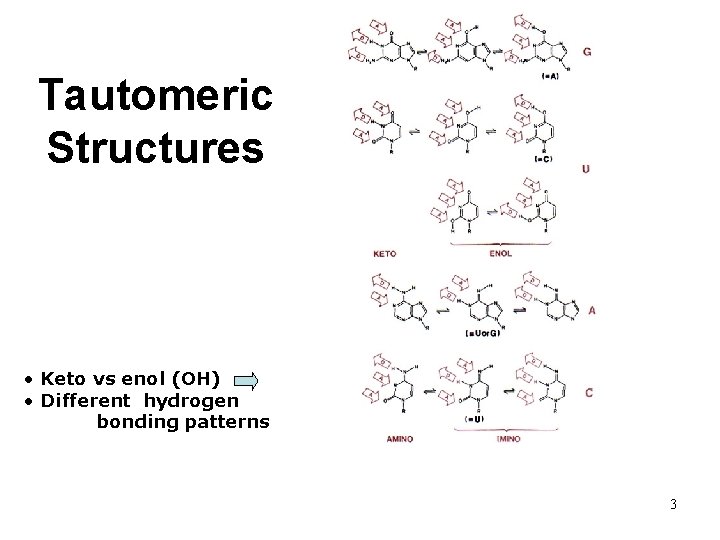
Tautomeric Structures • Keto vs enol (OH) • Different hydrogen bonding patterns 3
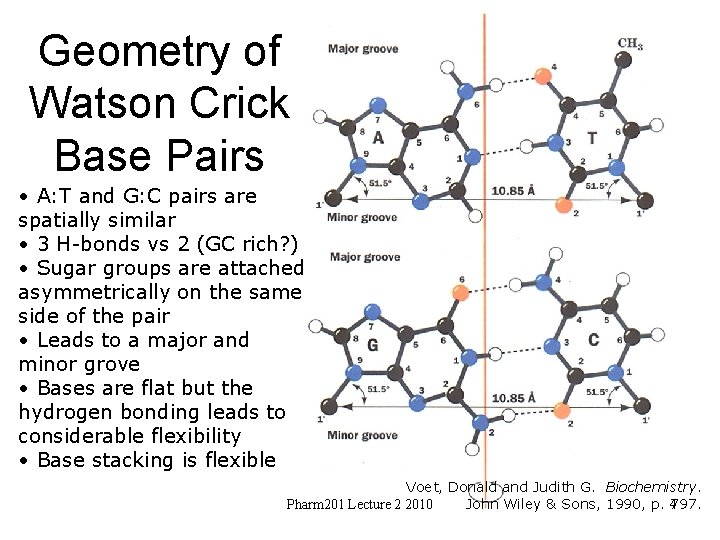
Geometry of Watson Crick Base Pairs • A: T and G: C pairs are spatially similar • 3 H-bonds vs 2 (GC rich? ) • Sugar groups are attached asymmetrically on the same side of the pair • Leads to a major and minor grove • Bases are flat but the hydrogen bonding leads to considerable flexibility • Base stacking is flexible Voet, Donald and Judith G. Biochemistry. Pharm 201 Lecture 2 2010 John Wiley & Sons, 1990, p. 4797.
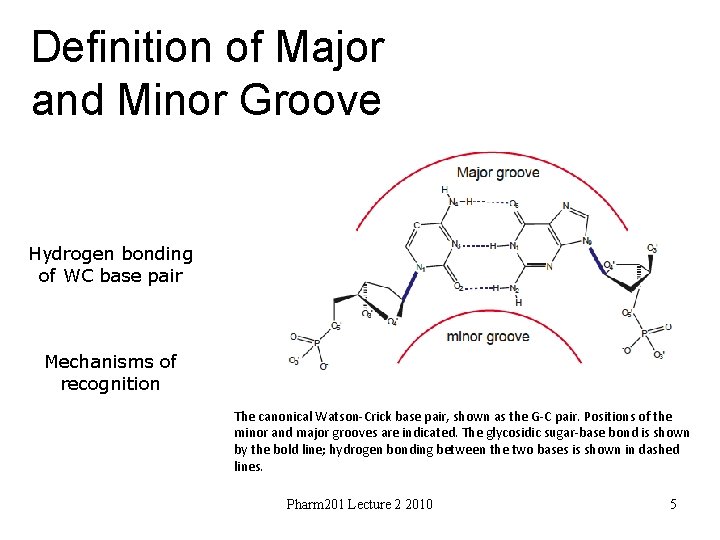
Definition of Major and Minor Groove Hydrogen bonding of WC base pair Mechanisms of recognition The canonical Watson-Crick base pair, shown as the G-C pair. Positions of the minor and major grooves are indicated. The glycosidic sugar-base bond is shown by the bold line; hydrogen bonding between the two bases is shown in dashed lines. Pharm 201 Lecture 2 2010 5
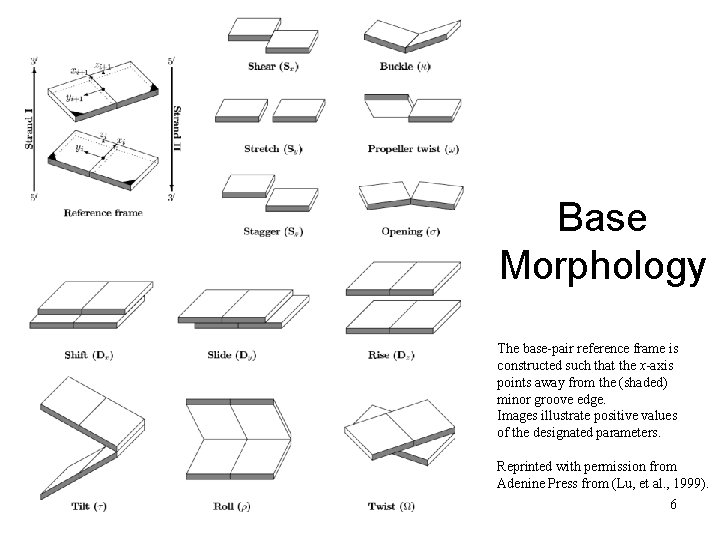
Base Morphology The base-pair reference frame is constructed such that the x-axis points away from the (shaded) minor groove edge. Images illustrate positive values of the designated parameters. Reprinted with permission from Adenine Press from (Lu, et al. , 1999). Pharm 201 Lecture 2 2010 6
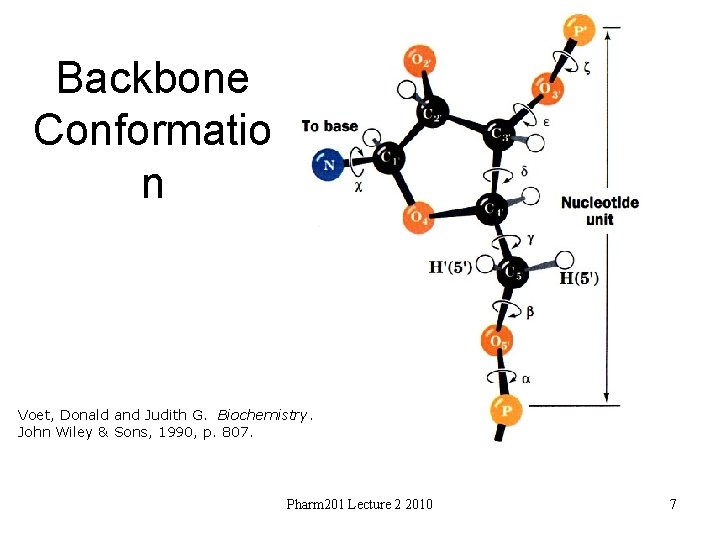
Backbone Conformatio n Voet, Donald and Judith G. Biochemistry. John Wiley & Sons, 1990, p. 807. Pharm 201 Lecture 2 2010 7
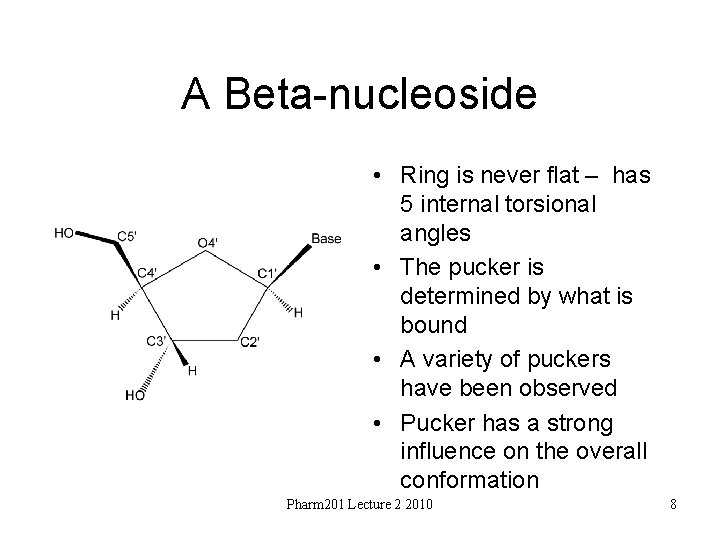
A Beta-nucleoside • Ring is never flat – has 5 internal torsional angles • The pucker is determined by what is bound • A variety of puckers have been observed • Pucker has a strong influence on the overall conformation Pharm 201 Lecture 2 2010 8
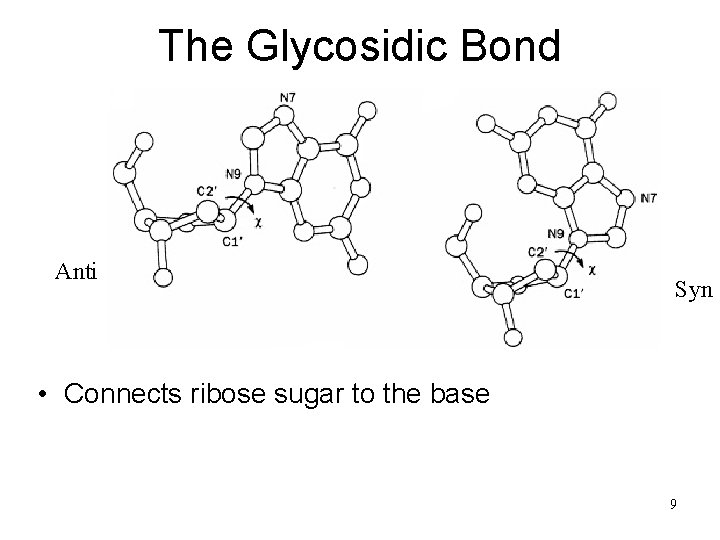
The Glycosidic Bond Anti Syn • Connects ribose sugar to the base 9
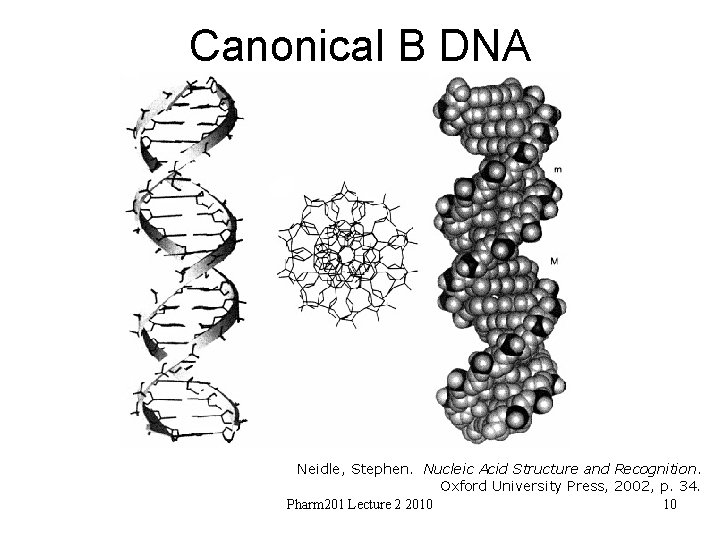
Canonical B DNA Neidle, Stephen. Nucleic Acid Structure and Recognition. Oxford University Press, 2002, p. 34. Pharm 201 Lecture 2 2010 10
Canonical B DNA • First determined experimentally by fiber diffraction (Arnott) • C 2’-endo sugar puckers • High anti glycosidic angles • Right handed – 10 base pairs per turn • Bases perpendicular to the helix axis and stacked over the axis • Overall bending as much as 15 degrees (result of base morphologies – twist and roll) – {machine learning – sequence vs overall conformation? } • Over 230 structures 25 with base mis-pairing – only cause local perturbations • Strong influence of hydration along spine http: //ndbserver. rutgers. edu/index. html Pharm 201 Lecture 2 2010 11
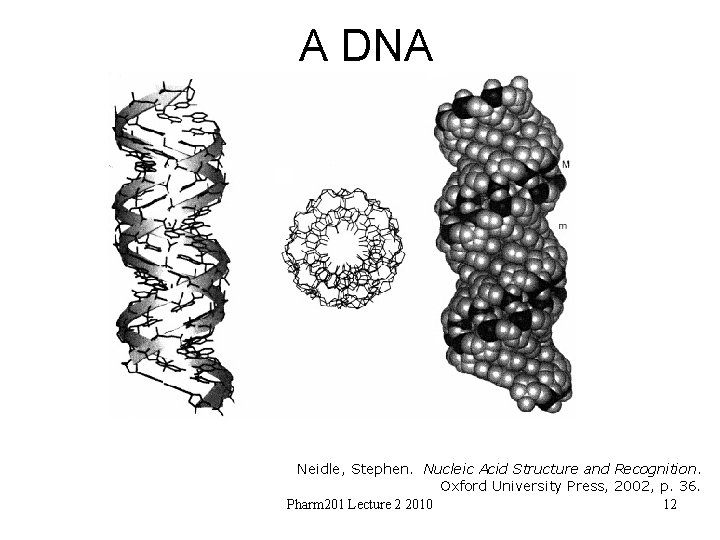
A DNA Neidle, Stephen. Nucleic Acid Structure and Recognition. Oxford University Press, 2002, p. 36. Pharm 201 Lecture 2 2010 12
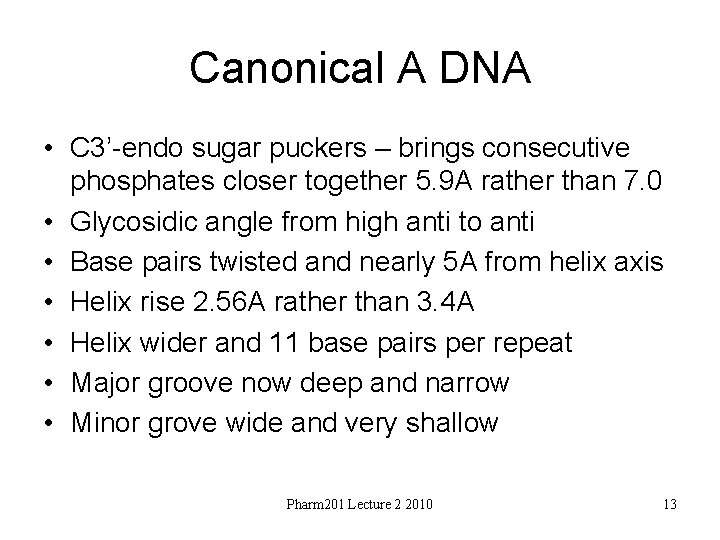
Canonical A DNA • C 3’-endo sugar puckers – brings consecutive phosphates closer together 5. 9 A rather than 7. 0 • Glycosidic angle from high anti to anti • Base pairs twisted and nearly 5 A from helix axis • Helix rise 2. 56 A rather than 3. 4 A • Helix wider and 11 base pairs per repeat • Major groove now deep and narrow • Minor grove wide and very shallow Pharm 201 Lecture 2 2010 13
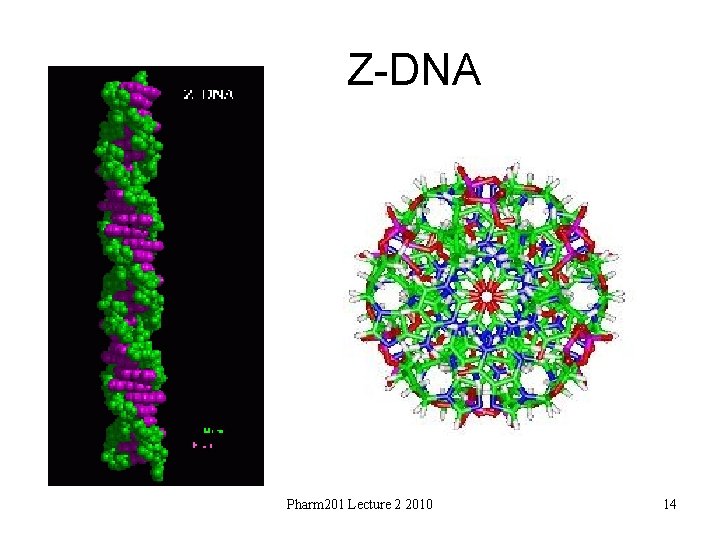
Z-DNA Pharm 201 Lecture 2 2010 14

Z-DNA • • • Helix has left-handed sense Can be formed in vivo, given proper sequence and superhelical tension, but function remains obscure. Narrower, more elongated helix than A or B. Major "groove" not really groove Narrow minor groove Conformation favored by high salt concentrations, some base substitutions, but requires alternating purine-pyrimidine sequence. N 2 -amino of G H-bonds to 5' PO: explains slow exchange of proton, need for G purine. Base pairs nearly perpendicular to helix axis Gp. C repeat, not single base-pair – P-P distances: vary for Gp. C and Cp. G – Gp. C stack: good base overlap – Cp. G: less overlap. • • Zigzag backbone due to C sugar conformation compensating for G glycosidic bond conformation Conformations: – G; syn, C 2'-endo – C; anti, C 3'-endo Pharm 201 Lecture 2 2010 15

Z-DNA • • Convex major groove Deep minor groove Alternate C then G Spine of hydration Pharm 201 Lecture 2 2010 16
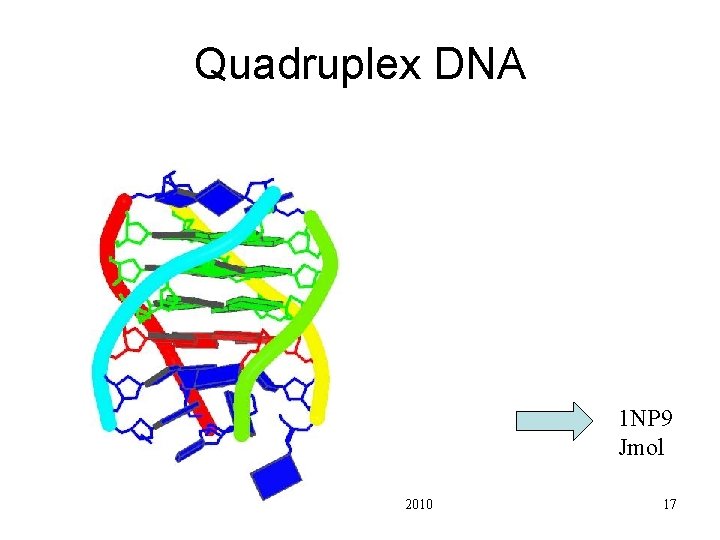
Quadruplex DNA 1 NP 9 Jmol Pharm 201 Lecture 2 2010 17
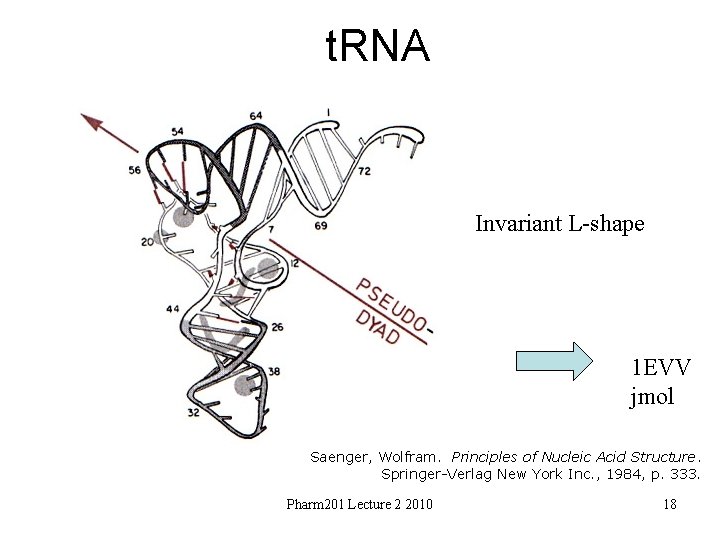
t. RNA Invariant L-shape 1 EVV jmol Saenger, Wolfram. Principles of Nucleic Acid Structure. Springer-Verlag New York Inc. , 1984, p. 333. Pharm 201 Lecture 2 2010 18
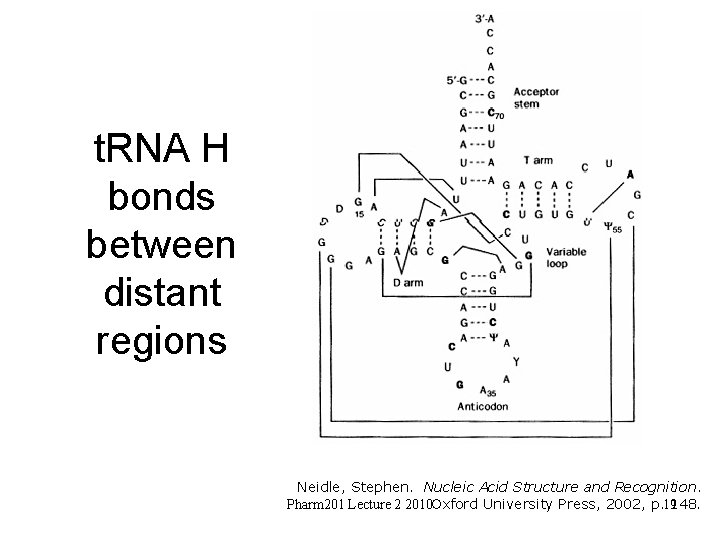
t. RNA H bonds between distant regions Neidle, Stephen. Nucleic Acid Structure and Recognition. Pharm 201 Lecture 2 2010 Oxford University Press, 2002, p. 19148.
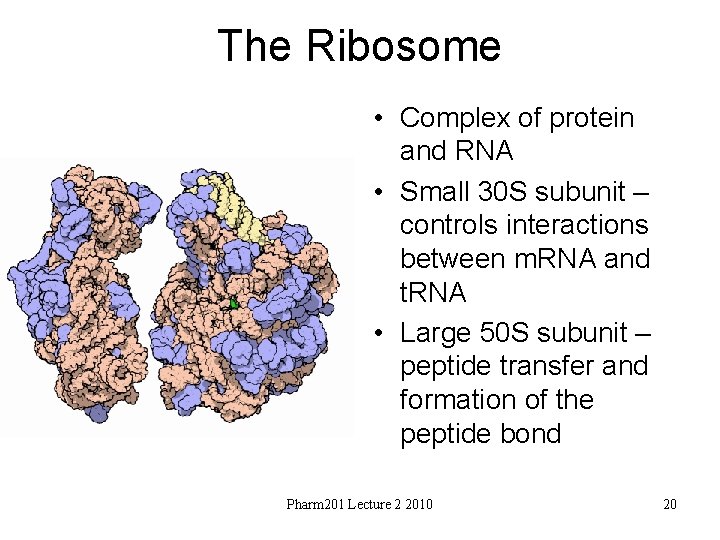
The Ribosome • Complex of protein and RNA • Small 30 S subunit – controls interactions between m. RNA and t. RNA • Large 50 S subunit – peptide transfer and formation of the peptide bond Pharm 201 Lecture 2 2010 20
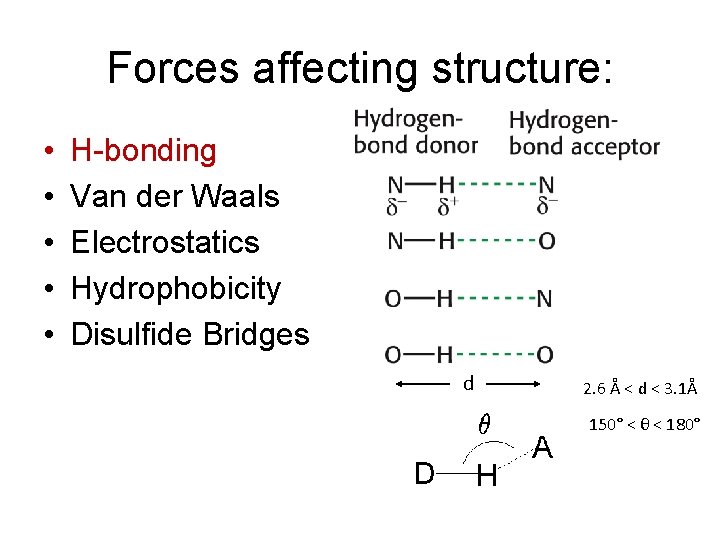
Forces affecting structure: • • • H-bonding Van der Waals Electrostatics Hydrophobicity Disulfide Bridges d 2. 6 Å < d < 3. 1Å 150° < θ < 180°
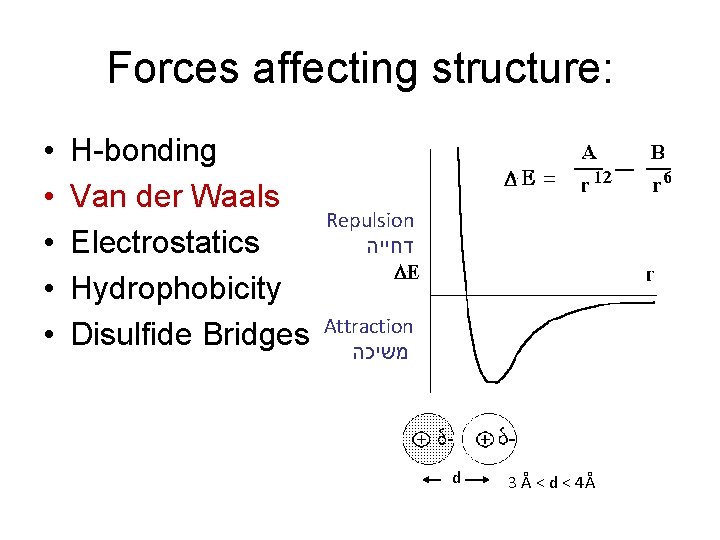
Forces affecting structure: • • • H-bonding Van der Waals Electrostatics Hydrophobicity Disulfide Bridges Repulsion דחייה Attraction משיכה d 3 Å < d < 4Å
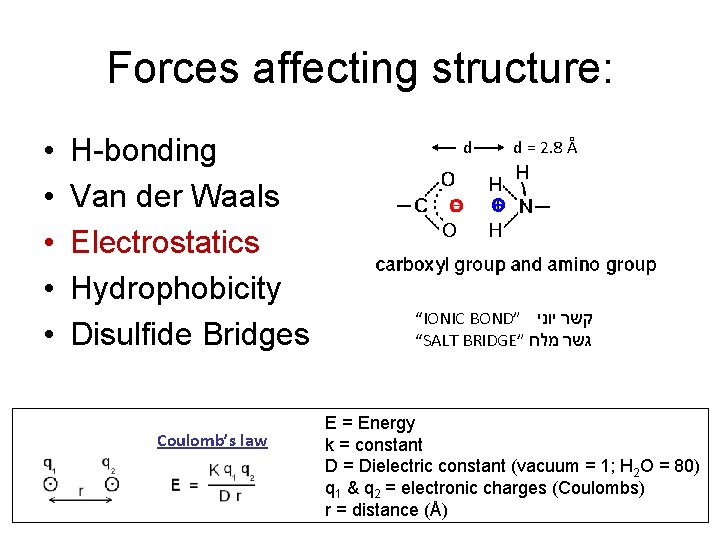
Forces affecting structure: • • • H-bonding Van der Waals Electrostatics Hydrophobicity Disulfide Bridges Coulomb’s law d d = 2. 8 Å “IONIC BOND” קשר יוני “SALT BRIDGE” גשר מלח E = Energy k = constant D = Dielectric constant (vacuum = 1; H 2 O = 80) q 1 & q 2 = electronic charges (Coulombs) r = distance (Å)
Forces affecting structure: • • • H-bonding Van der Waals Electrostatics Hydrophobicity Disulfide Bridges
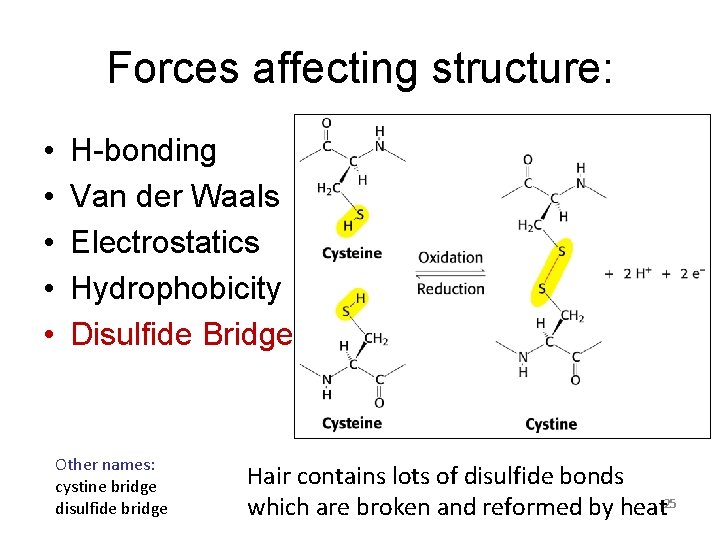
Forces affecting structure: • • • H-bonding Van der Waals Electrostatics Hydrophobicity Disulfide Bridges Other names: cystine bridge disulfide bridge Hair contains lots of disulfide bonds which are broken and reformed by heat 25
Structure Representation and Coordinates Format Lecture 3 Structural Bioinformatics Dr. Avraham Samson 81 -871
The PDB Format • A full description is here • It was designed around an 80 column punched card! • It was designed to be human readable • It is used by almost every piece of software that deals with structural data 27
The PDB Format - Records • Every PDB file may be broken into a number of lines terminated by an end-of-line indicator. Each line in the PDB entry file consists of 80 columns. The last character in each PDB entry should be an end-of-line indicator. • Each line in the PDB file is self-identifying. The first six columns of every line contain a record name, left-justified and blank-filled. This must be an exact match to one of the stated record names. • The PDB file may also be viewed as a collection of record types. Each record type consists of one or more lines. • Each record type is further divided into fields. 28
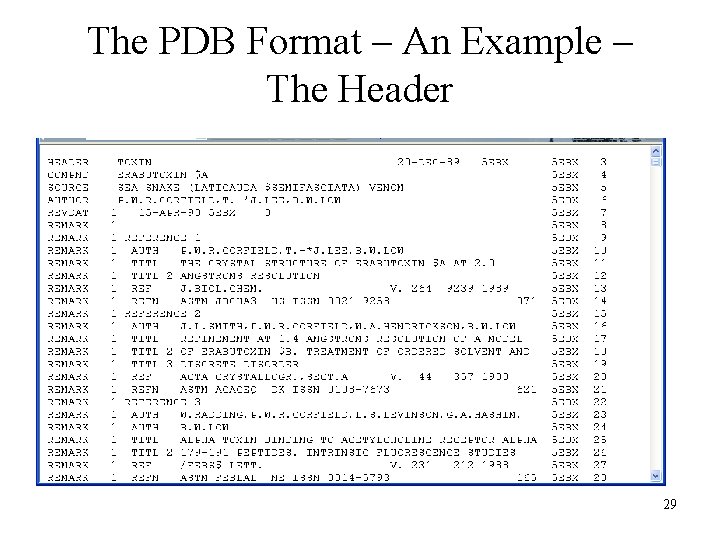
The PDB Format – An Example – The Header 29
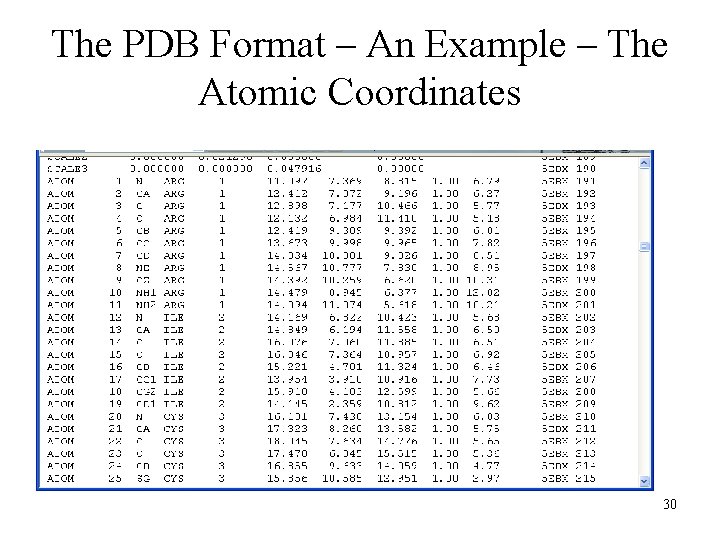
The PDB Format – An Example – The Atomic Coordinates 30

The Description – Atom Records 31

What is Wrong with this Approach? • The description and the data are separate • Parsing is a nightmare – the most complex piece of code we have in our research laboratory probably remains the PDB parser • There are no relationships between items of data • Some data just cannot be parsed • The fixed column format cannot represent some of today’s structures … 32

Structures are Spread Over Multiple Files – Most Users are Not Aware of this 33
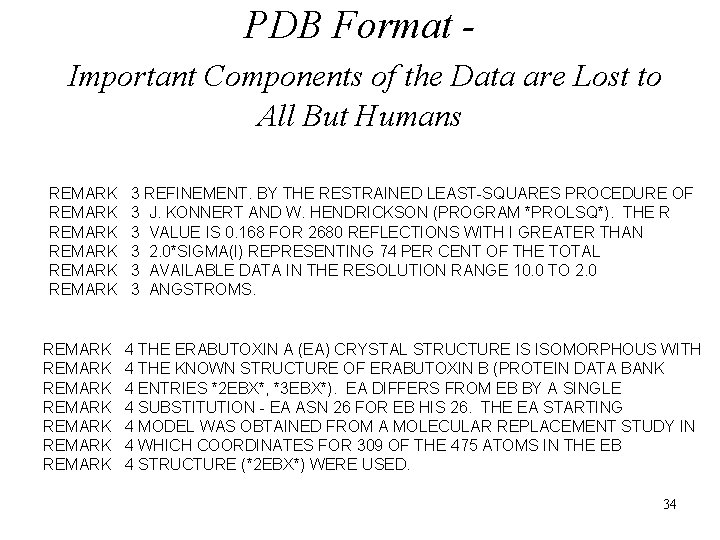
PDB Format Important Components of the Data are Lost to All But Humans REMARK REMARK REMARK REMARK 3 REFINEMENT. BY THE RESTRAINED LEAST-SQUARES PROCEDURE OF 3 J. KONNERT AND W. HENDRICKSON (PROGRAM *PROLSQ*). THE R 3 VALUE IS 0. 168 FOR 2680 REFLECTIONS WITH I GREATER THAN 3 2. 0*SIGMA(I) REPRESENTING 74 PER CENT OF THE TOTAL 3 AVAILABLE DATA IN THE RESOLUTION RANGE 10. 0 TO 2. 0 3 ANGSTROMS. 4 THE ERABUTOXIN A (EA) CRYSTAL STRUCTURE IS ISOMORPHOUS WITH 4 THE KNOWN STRUCTURE OF ERABUTOXIN B (PROTEIN DATA BANK 4 ENTRIES *2 EBX*, *3 EBX*). EA DIFFERS FROM EB BY A SINGLE 4 SUBSTITUTION - EA ASN 26 FOR EB HIS 26. THE EA STARTING 4 MODEL WAS OBTAINED FROM A MOLECULAR REPLACEMENT STUDY IN 4 WHICH COORDINATES FOR 309 OF THE 475 ATOMS IN THE EB 4 STRUCTURE (*2 EBX*) WERE USED. 34

mm. CIF Was Developed to Address these Problems Methods in Enzymology. 1997 277, 571 -590 35

mm. CIF – Scope of the Initial Effort • All PDB data should be captured • Describe a paper’s material and methods section • Describe biologically active molecule • Fully describe secondary structure but not tertiary or quaternary • Describe details of chemistry (inc. 2 D) • Meaningful 3 D views 36

mm. CIF - Extract from a Data File loop_ _atom_site. group_PDB _atom_site. type_symbol _atom_site. label_atom_id _atom_site. label_comp_id _atom_site. label_asym_id _atom_site. label_seq_id _atom_site. label_alt_id _atom_site. Cartn_x _atom_site. Cartn_y _atom_site. Cartn_z _atom_site. occupancy _atom_site. B_iso_or_equiv _atom_site. footnote_id _atom_site. entity_seq_num _atom_site. id ATOM N N VAL A 11. 25. 360 30. 691 11. 795 1. 00 17. 93. 1 11 1 ATOM C CA VAL A 11. 25. 970 31. 965 12. 332 1. 00 17. 75. 1 11 2 ATOM C C VAL A 11. 25. 569 32. 010 13. 881 1. 00 17. 83. 1 11 3 37
Summary • mm. CIF has provided the PDB with a robust data representation which serves as conceptual and physical schema upon which the current RCSB, PDBe and PDBj are built • This work predated XML and XML-schema but embodies the important concepts inherent in these descriptions • mm. CIF was later exactly converted into XML and is now used more than mm. CIF, but much less than the old PDB format • Today mm. CIF has no advantage over PDB 38

Other representations • SMILES http: //en. wikipedia. org/wiki/Simplified_mol ecular-input_line-entry_system 39

Other representations 40

Representing Positions • Cartesian coordinates (x, y, z) are an easy and natural means of representing a position in 3 D space • There are many other alternatives such as polar notation (r, θ, φ) and you can invent others if you want to
Other representations -Cartesian coordinates vs. polar coordinates 42
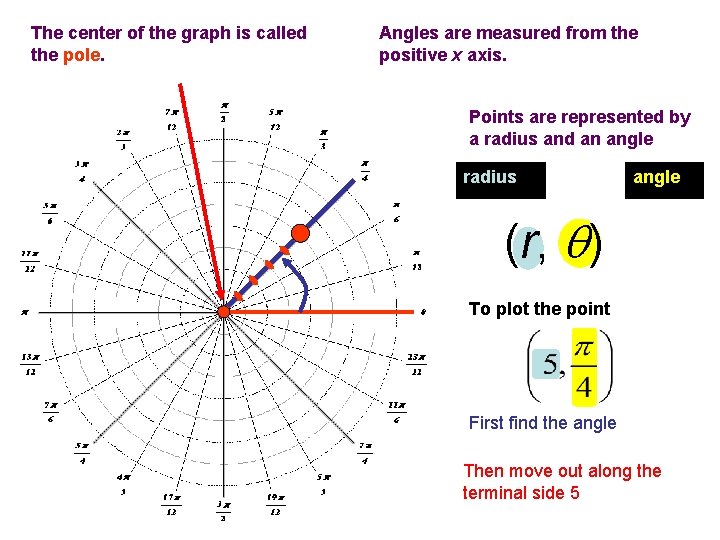
The center of the graph is called the pole. Angles are measured from the positive x axis. Points are represented by a radius and an angle radius angle (r, ) To plot the point First find the angle Then move out along the terminal side 5
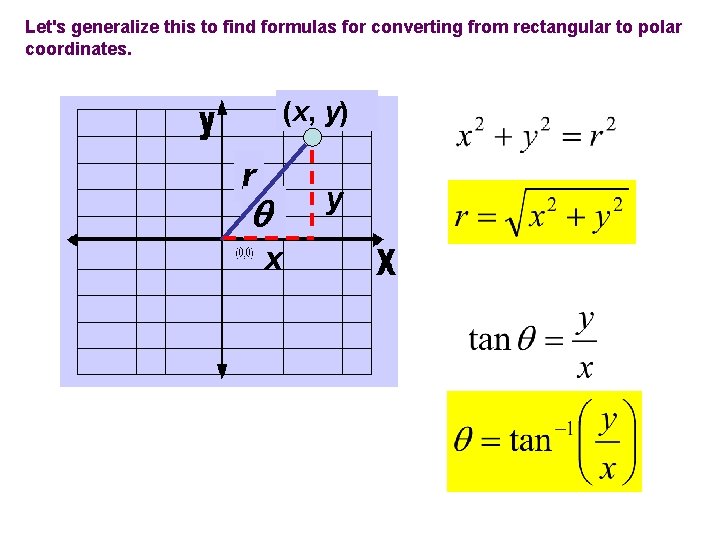
Let's generalize this to find formulas for converting from rectangular to polar coordinates. (x, y) r x y
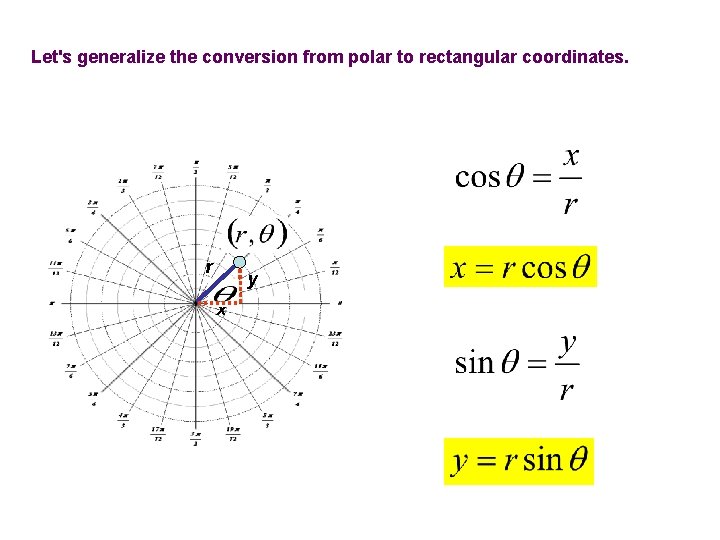
Let's generalize the conversion from polar to rectangular coordinates. r y x

• How would you calculate distance? • How would you calculate centroid? • How would you calculate dihedral angle? 46
- Slides: 46