Diversity of Life Introduction to Biological Classification Bio

Diversity of Life: Introduction to Biological Classification Bio. Ed. Online

Why Do We Classify Organisms? n n Biologists group organisms to represent similarities and proposed relationships. Classification systems change with expanding knowledge about new and well-known organisms. Tacitus bellus Bio. Ed Online www. Bio. Ed. Online. org

Classification n Binomial Nomenclature n n Hierarchical Classification n n Two part name (Genus, species) Seven Taxonomic Catagroies Systematics n Study of the evolution of biological diversity Leucaena leucocephala Lead tree Bio. Ed Online www. Bio. Ed. Online. org

Binomial Nomenclature n n Carolus von Linnaeus Two-word naming system n Genus n n Noun, Capitalized, Underlined or Italicized Species n Descriptive, Lower Case, Underlined or Italicized Carolus von Linnaeus (1707 -1778) Swedish scientist who laid the foundation for modern taxonomy Bio. Ed Online www. Bio. Ed. Online. org
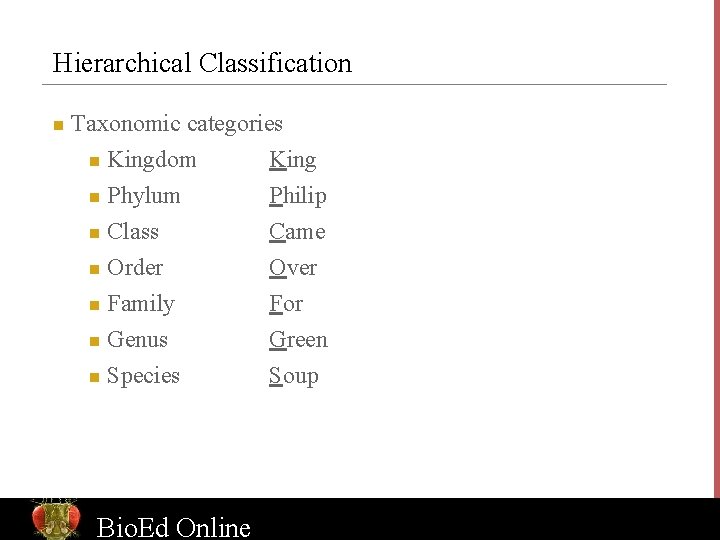
Hierarchical Classification n Taxonomic categories Kingdom n Phylum n Class n Order n Family n Genus King Philip Came Over For Green Species Soup n n Bio. Ed Online www. Bio. Ed. Online. org
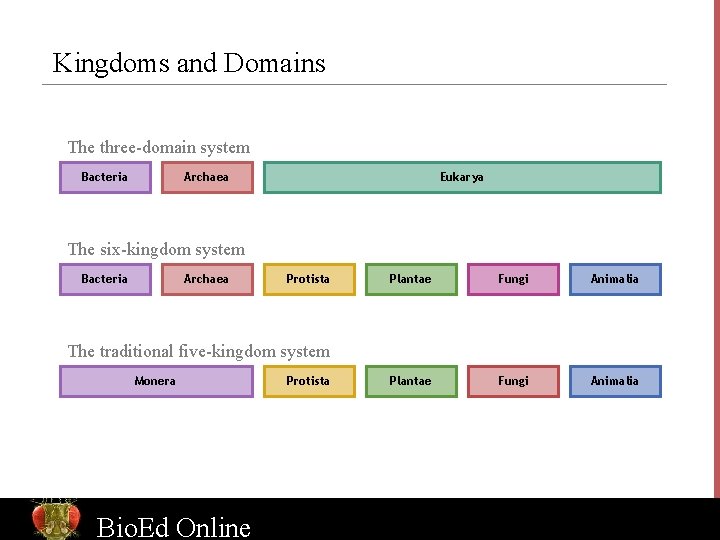
Kingdoms and Domains The three-domain system Bacteria Archaea Eukarya The six-kingdom system Bacteria Archaea Protista Plantae Fungi Animalia The traditional five-kingdom system Monera Bio. Ed Online Protista www. Bio. Ed. Online. org
Systematics: Evolutionary Classification of Organisms n Systematics is the study of the evolution of biological diversity, and combines data from the following areas. n Fossil record n Comparative homologies Cladistics n Comparative sequencing of DNA/RNA among organisms n Molecular clocks n Bio. Ed Online www. Bio. Ed. Online. org
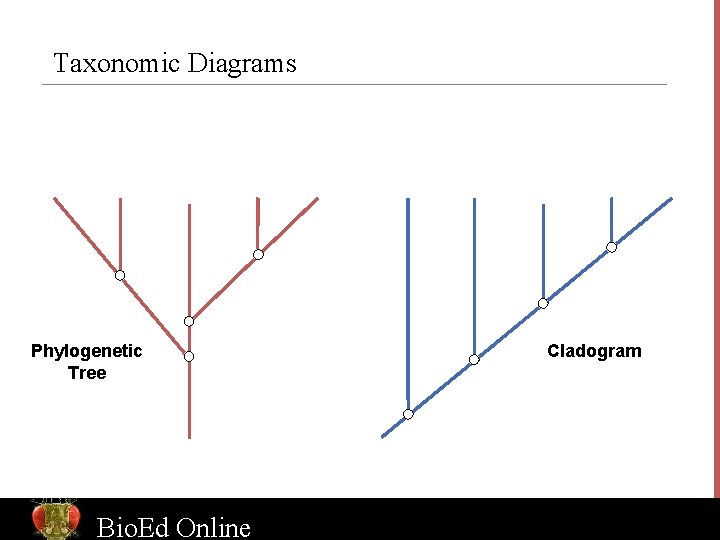
Taxonomic Diagrams Phylogenetic Tree Bio. Ed Online Cladogram www. Bio. Ed. Online. org
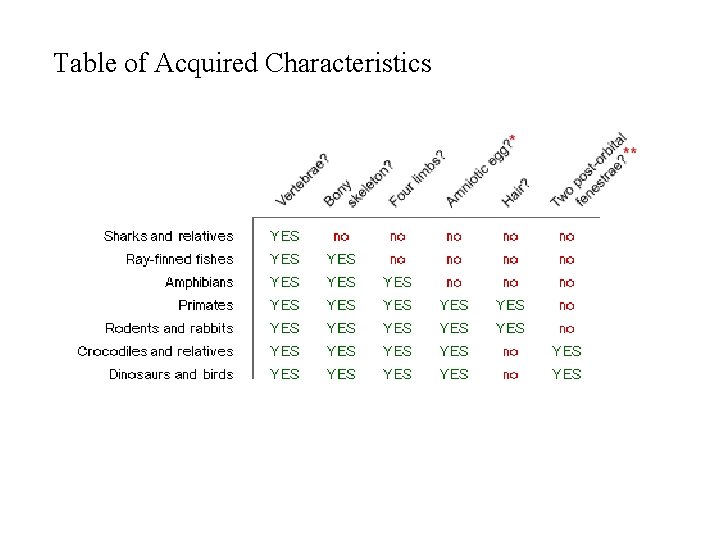
Table of Acquired Characteristics

Now it’s your turn to make a tree! n Animals n Drosophila melanogaster (fruit fly) n Homo sapiens (you) n Heterodontus francisci (horn shark) n Chelonia mydas (green sea turtle) n Xenopus laevis (african clawed frog) n Gallus gallus (chicken) n Mus musculus (house mouse) n Danio rerio (zebrafish) n Naja naja (Indian Cobra) n Rousettus aegyptiacus (Egyptian fruit bat) n Caenorhabditis elegans (roundworm nematode) n Aurelia aurita (jellyfish) n Asterias amurensis (northern Pacific sea star) n Dosidicus gigas (giant squid) n Saccharomyces cerevisiae (baker’s yeast) n Macropus robustus (Eastern Wallaroo) n Characters n Homeothermy n n Appendages n n n Endo, exo, neither Amniotic sac Body plan n n Smooth, scales, hair, feathers Skeleton n n Fins, legs, wings Skin n n Warm vs cold blooded Symmetry, # limbs, etc Can you think of others?
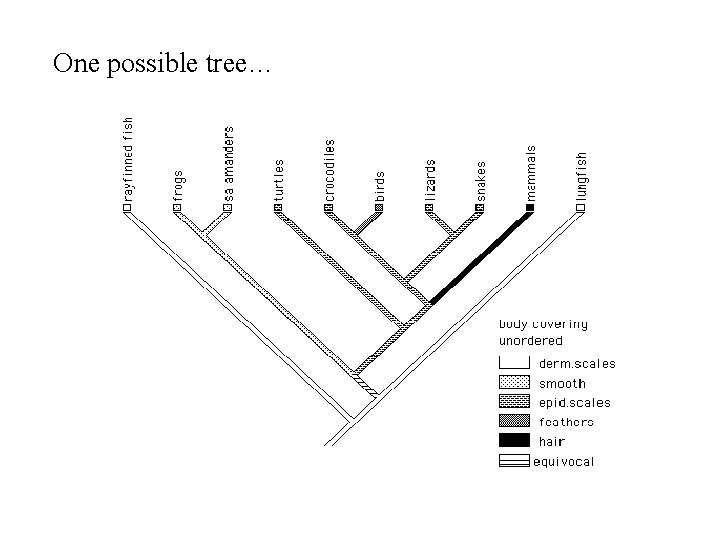
One possible tree…

Taxonomic Diagrams Mammals Turtles Lizards and Snakes Crocodiles Phylogenetic Tree Bio. Ed Online Birds Mammals Turtles Lizards and Snakes Crocodiles Cladogram Birds

Tree of Life (cladogram style) Yeast Jellyfish Squid Nematode Fruit fly Starfish Sharks Bony Fish Amphibians Snakes Birds Turtles Kangaroo Mice Bats Humans
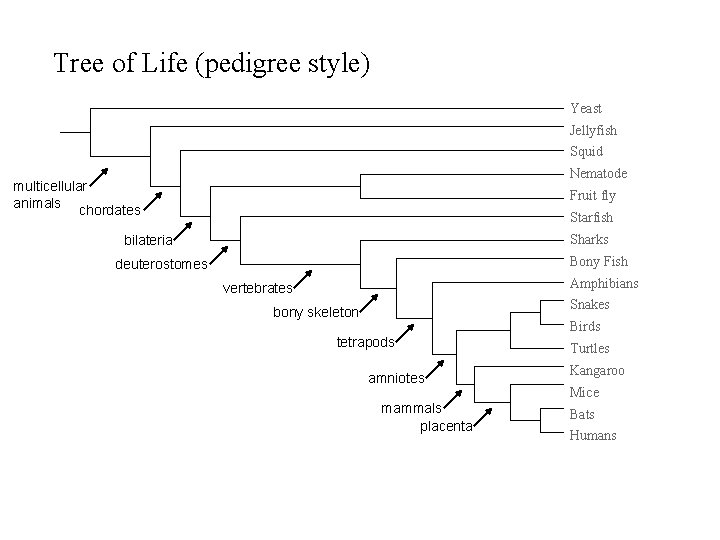
Tree of Life (pedigree style) Yeast Jellyfish Squid Nematode multicellular animals chordates Fruit fly Starfish Sharks bilateria Bony Fish deuterostomes Amphibians vertebrates Snakes bony skeleton Birds tetrapods amniotes mammals placenta Turtles Kangaroo Mice Bats Humans
Phylogenetics! n n Open your laptops Go to http: //www. tolweb. org/ n Learn about phylogenetics

Phylogenetics and Bioinformatics! n n Open your laptops Go to http: //www. ncbi. nlm. nih. gov/ n Tour the website n Learn about bioinformatics n Download two files from Study Wiz n n Species accession numbers. txt Cyt. B sequences. txt

Protein BLAST Instructions n n n Go To BLAST website n http: //www. ncbi. nlm. nih. gov/blast/ Click on “protein blast” Enter the accession number for human cytochrome B protein (NP_536855. 1) Paste the other cytochrome B accession numbers into the “Entrez Query” box n This will limit our comparison to only the mitochondrial records we specify here Click BLAST! Click on “Distance tree of results” n Select “Taxonomic Name” under “Sequence Label” n Select the “Slanted” tab to view a cladogram style

Cyt. B BLAST Tree (local alignment)

Multiple Sequence Alignment n Go to align. genome. jp n Copy and paste the Cytochrome B sequences n Make a dendrogram with distances n Answer questions on sheet
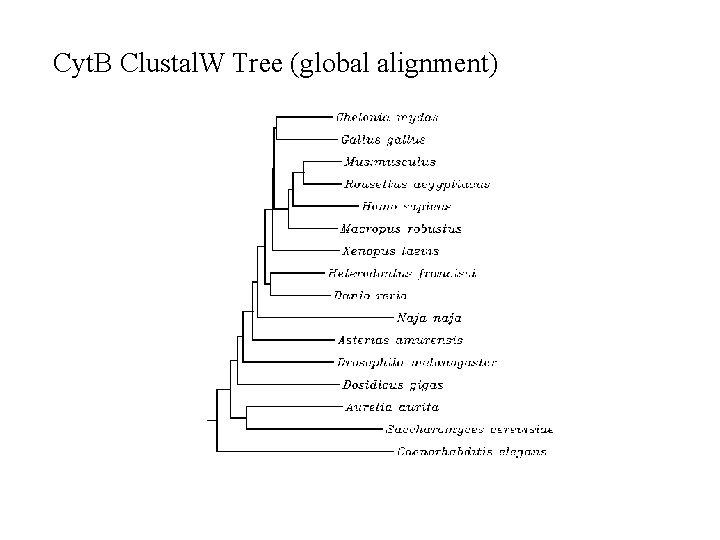
Cyt. B Clustal. W Tree (global alignment)
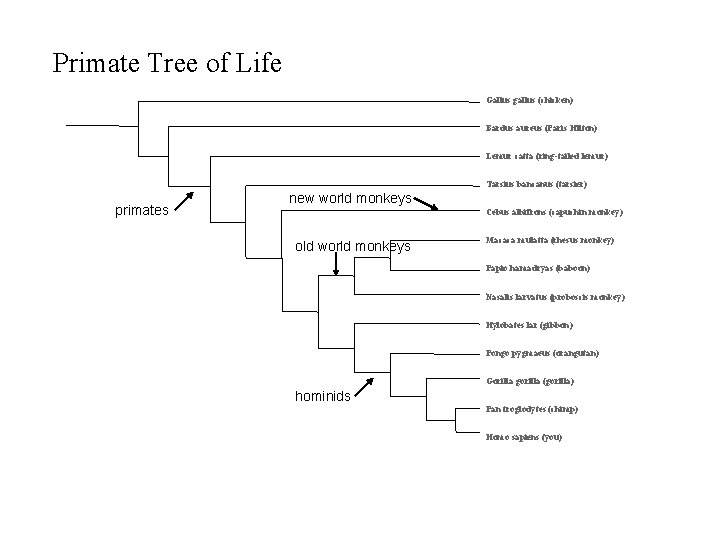
Primate Tree of Life Gallus gallus (chicken) Bardus aureus (Paris Hilton) Lemur catta (ring-tailed lemur) Tarsius bancanus (tarsier) primates new world monkeys Cebus albifrons (capuchin monkey) old world monkeys Macaca mulatta (rhesus monkey) Papio hamadryas (baboon) Nasalis larvatus (proboscis monkey) Hylobates lar (gibbon) Pongo pygmaeus (orangutan) Gorilla gorilla (gorilla) hominids Pan troglodytes (chimp) Homo sapiens (you)

Nucleotide BLAST Instructions n Go To BLAST website n http: //www. ncbi. nlm. nih. gov/blast/ n Click on “nucleotide blast” n Enter the accession number for human mitochondrial genome (NC_001807) n n Select database “Reference genomic sequences” n This will make sure we search within only genomic records Paste the other mitochondrial accession numbers into the “Entrez Query” box n This will limit our comparison to only the mitochondrial records we specify here Click BLAST! Click on “Distance tree of results” n Select “Taxonomic Name” under “Sequence Label” n Select the “Slanted” tab to view a cladogram style Bio. Ed Online www. Bio. Ed. Online. org
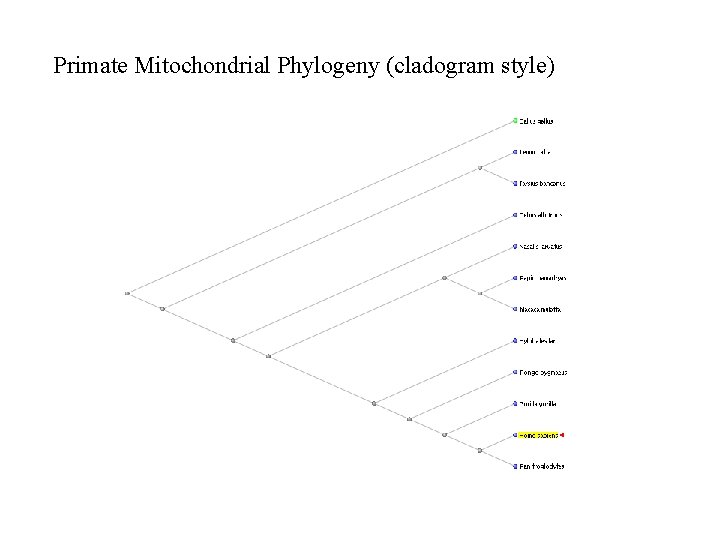
Primate Mitochondrial Phylogeny (cladogram style)
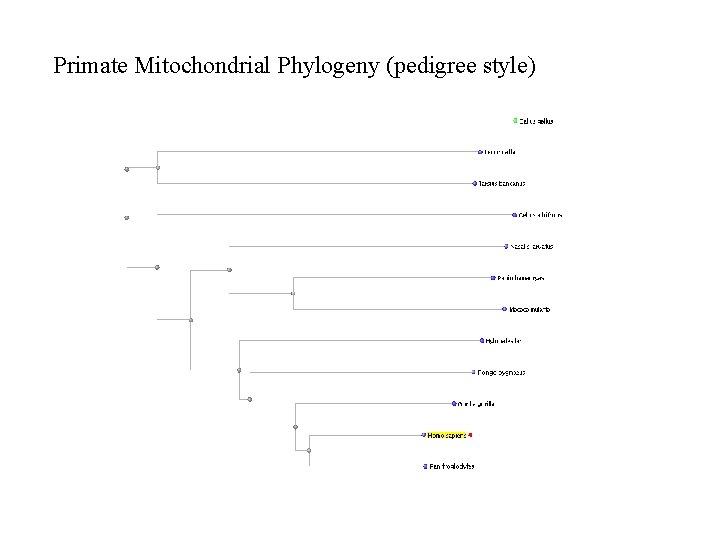
Primate Mitochondrial Phylogeny (pedigree style)
- Slides: 24