Distance methods Distance methods UPGMA similar to hierarchical








- Slides: 14

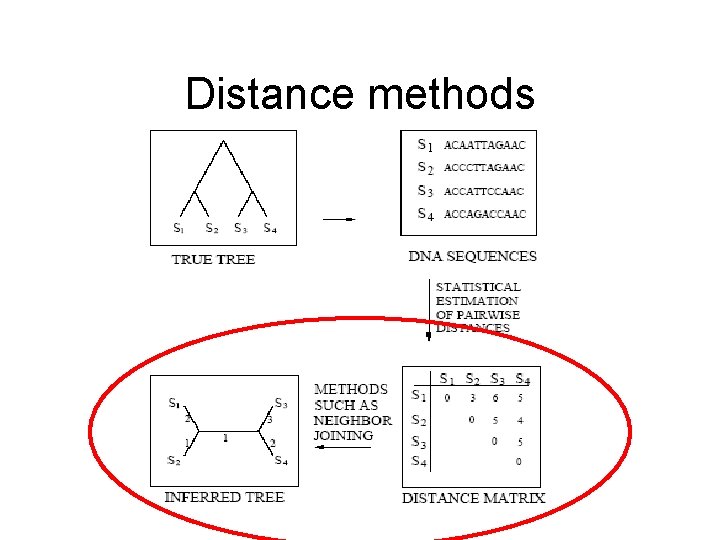
Distance methods

Distance methods • UPGMA: similar to hierarchical clustering but not additive • Neighbor-joining: more sophisticated and additive • What is additivity?

Additivity

UPGMA is not additive but works for ultrametric trees. Takes O(n^3) time A 10 B 3 A 3 3 B C 3 C D D B C 6 26 26 D 26 26 6

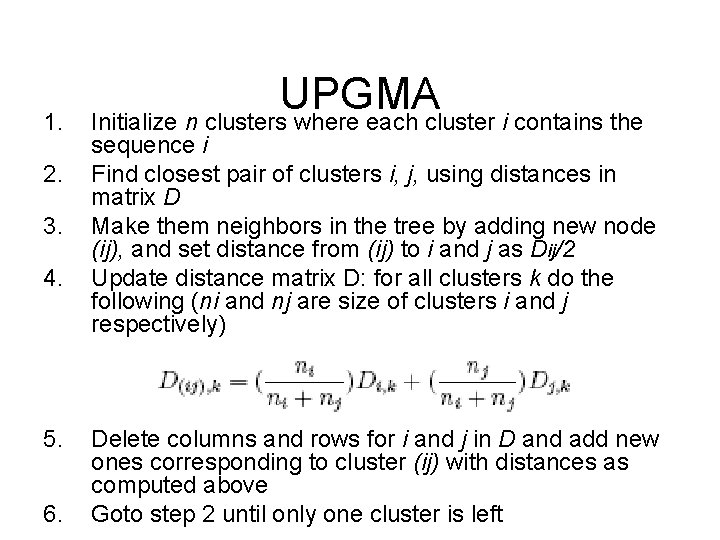
1. 2. 3. 4. 5. 6. UPGMA Initialize n clusters where each cluster i contains the sequence i Find closest pair of clusters i, j, using distances in matrix D Make them neighbors in the tree by adding new node (ij), and set distance from (ij) to i and j as Dij/2 Update distance matrix D: for all clusters k do the following (ni and nj are size of clusters i and j respectively) Delete columns and rows for i and j in D and add new ones corresponding to cluster (ij) with distances as computed above Goto step 2 until only one cluster is left

UPGMA A A B C D B C 6 32 32 D 13 13 32 32 6 3 A 3 3 B C 3 D

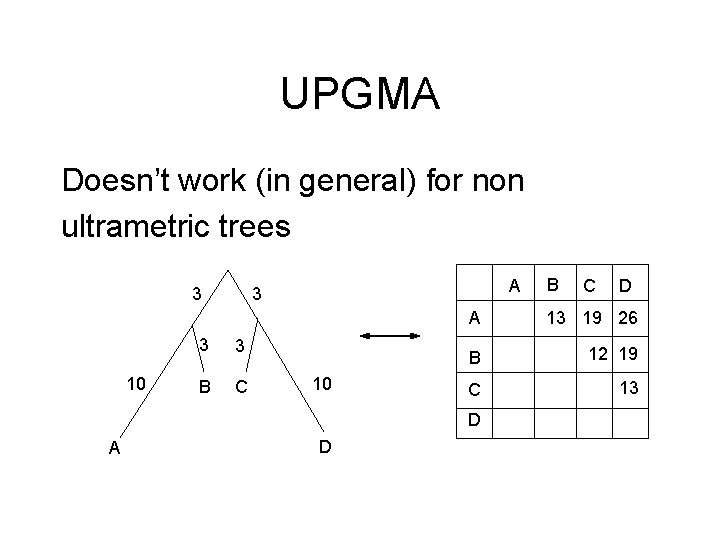
UPGMA Doesn’t work (in general) for non ultrametric trees 3 10 A 3 3 3 B C 10 D C D A 13 19 26 B 12 19 C 13 D A B

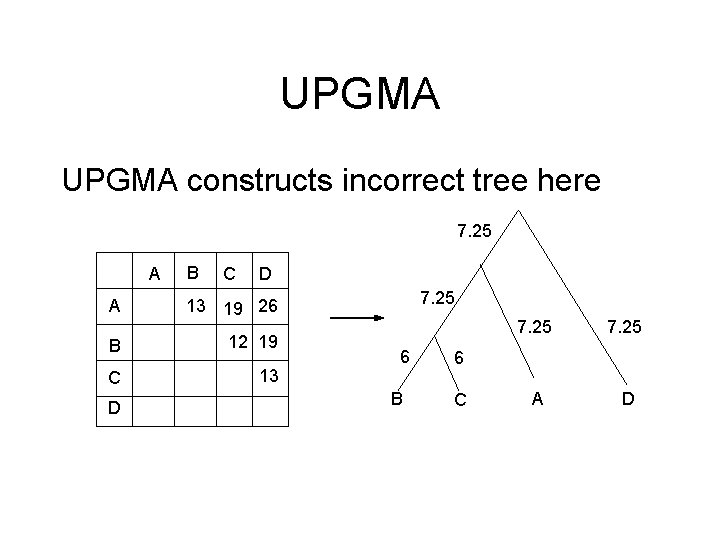
UPGMA constructs incorrect tree here 7. 25 A B C D A 13 19 26 B 12 19 C 13 D 7. 25 6 B 7. 25 A D 6 C

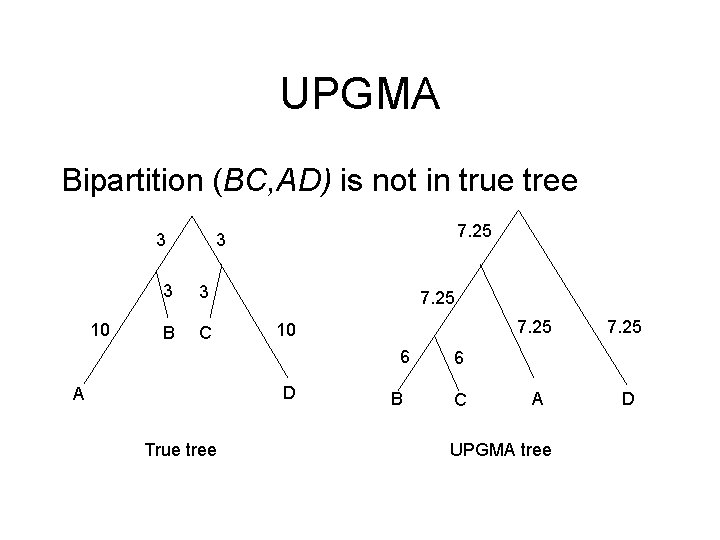
UPGMA Bipartition (BC, AD) is not in true tree 3 10 7. 25 3 3 3 B C 7. 25 10 6 D A True tree B 7. 25 A D 6 C UPGMA tree

Neighbor joining 1. 2. 3. 4. Additive and O(n^3) time Initialization: same as UPGMA For each species compute Select i and j for which is minimum 5. Make them neighbors in the tree by adding new node (ij), and set distance from (ij) to i and j as

Neighbor joining 6. Update distance matrix D: for all clusters k do the following 7. Delete columns and rows for i and j in D and add new ones corresponding to cluster (ij) with distances as computed above 8. Go to 3 until two nodes/clusters are left

NJ NJ constructs the correct tree for additive matrices 3 10 A 3 3 3 B C 10 D C D A 13 19 26 B 12 19 C 13 D A B

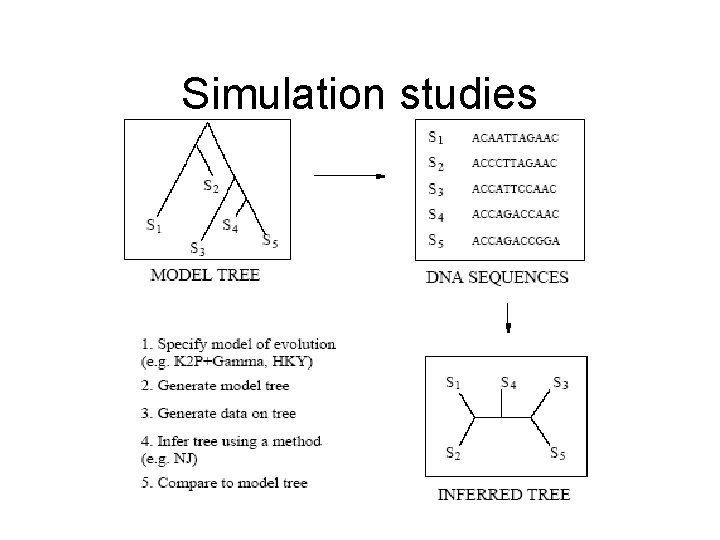
Simulation studies

Simulation studies • The true evolutionary tree is never known in practice. Simulation allows us to study accuracy of methods under biologically realistic scenarios • Mathematics behind the phylogenetics is often complex and challenging. Simulation allows us to study algorithms when not possible theoretically and also examine algorithm performance under various conditions such as different evolutionary rates, sequence lengths, or numbers of taxa
 Upgma tree generator
Upgma tree generator Upgma
Upgma Similar disuelve a similar
Similar disuelve a similar Unidad de medida de solubilidad
Unidad de medida de solubilidad Similar
Similar Paraphrasing proverbs
Paraphrasing proverbs A-wax pattern recognition
A-wax pattern recognition Types of ground in ncc
Types of ground in ncc How to solve for displacement
How to solve for displacement The ratio of input distance to output distance
The ratio of input distance to output distance Complete link clustering
Complete link clustering Occupational prestige ratings
Occupational prestige ratings Contagious diffusion
Contagious diffusion Pedro coelhoso
Pedro coelhoso Hierarchical temporal memory
Hierarchical temporal memory