Distance and Routing Labeling Schemes in Graphs with
Distance and Routing Labeling Schemes in Graphs with Applications in Cellular and Sensor Networks Feodor F. Dragan Computer Science Department Kent State University http: //www. cs. kent. edu/~dragan

Talk is based on • Known results for trees [Kannan&Naor&Rudich’ 88, Peleg’ 99 and ’ 00, Thorup&Zwick’ 01, Fraigniaud&Gavoille’ 01] • Two recent (Fall 2003) papers (by V. D. Chepoi (U. Marseille), F. F. Dragan and Yann Vaxes (U. Marseille) – Distance and Routing Labeling Schemes for Non. Positively Curved Plane Graphs (subm. to J. of Algorithms); – Addressing, Distances and Routing in Triangular Systems with Applications in Cellular and Sensor Networks (to appear in Proc of 4 th WNAN 04) We start with an overview on labeling schemes …
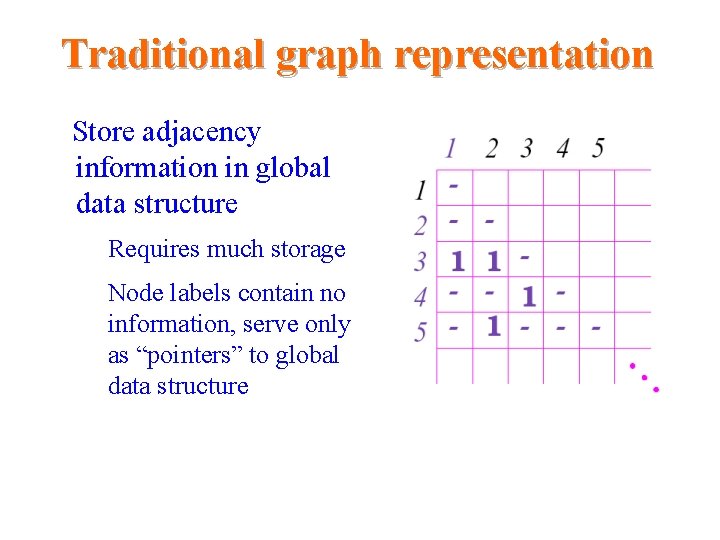
Traditional graph representation Store adjacency information in global data structure – Requires much storage – Node labels contain no information, serve only as “pointers” to global data structure
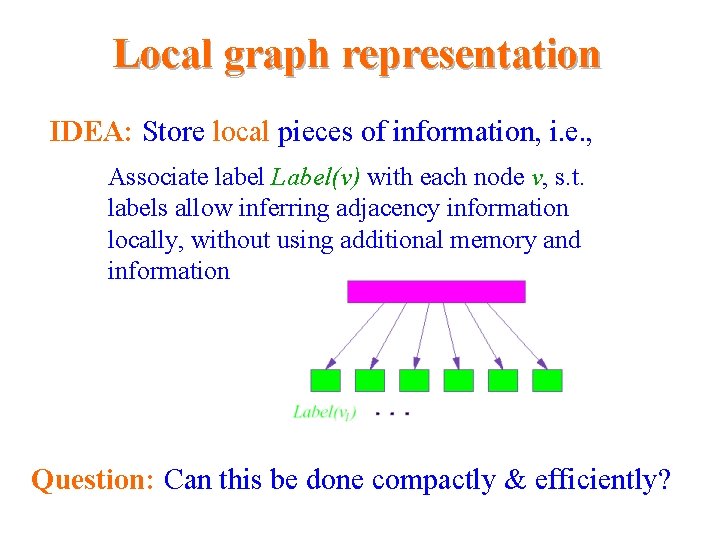
Local graph representation IDEA: Store local pieces of information, i. e. , – Associate label Label(v) with each node v, s. t. labels allow inferring adjacency information locally, without using additional memory and information Question: Can this be done compactly & efficiently?

“Ancient” Example: Hamming adjacency labeling Goal: Label each node v with m-bit label Label(v) s. t. Label(v), Label (u) allow deciding if u and v are adjacent: For any n-node graph there exists a Hamming distance adjacency labeling with m=2 n. D, T=4 D -4, where D = max node degree. [Breuer&Folkman’ 67]: Question: Can this be done with short labels?
![Interest Revived: Adjacency Labeling [Kannan&Naor&Rudich’ 88] What’s in label? Adjacency-labeling (for graph family F Interest Revived: Adjacency Labeling [Kannan&Naor&Rudich’ 88] What’s in label? Adjacency-labeling (for graph family F](http://slidetodoc.com/presentation_image/1a50e3b647fe44c84dad3dc1d834cb50/image-6.jpg)
Interest Revived: Adjacency Labeling [Kannan&Naor&Rudich’ 88] What’s in label? Adjacency-labeling (for graph family F ): 1. Function Label labeling nodes of any graph in F with short distinct labels (compactness) 2. Efficient algorithm for deciding adjacency of two nodes given their labels (efficiency) (polylogarithmic labels polylogarithmic time) Note: - Algorithm knows nothing beyond the labels (even, it does not know which graph they come from)
![Adjacency Labeling on Trees [Kannan&Naor&Rudich’ 88] Labeling: 1. Arbitrarily root the tree and prelabel Adjacency Labeling on Trees [Kannan&Naor&Rudich’ 88] Labeling: 1. Arbitrarily root the tree and prelabel](http://slidetodoc.com/presentation_image/1a50e3b647fe44c84dad3dc1d834cb50/image-7.jpg)
Adjacency Labeling on Trees [Kannan&Naor&Rudich’ 88] Labeling: 1. Arbitrarily root the tree and prelabel each node v with distinct integer I(v) from [1. . n]. 2. Label root r by Label(r)=(I(r)). 3. Label each non-root node v with parent w by Label(v)=(I(v), I(w)). Labels contain bits
![Adjacency Labeling on Trees [Kannan&Naor&Rudich’ 88] Labeling: (with bits) 1. Arbitrarily root the tree Adjacency Labeling on Trees [Kannan&Naor&Rudich’ 88] Labeling: (with bits) 1. Arbitrarily root the tree](http://slidetodoc.com/presentation_image/1a50e3b647fe44c84dad3dc1d834cb50/image-8.jpg)
Adjacency Labeling on Trees [Kannan&Naor&Rudich’ 88] Labeling: (with bits) 1. Arbitrarily root the tree and prelabel each node v with distinct integer I(v) from [1. . n]. 2. Label root r by Label(r)=(I(r)). 3. Label each non-root node v with parent w by Label(v)=(I(v), I(w)). Adjacency Decoder for u, v: Check if 1 st entry in Label(v) = 2 nd entry in Label(u) or vice versa. Constant time!
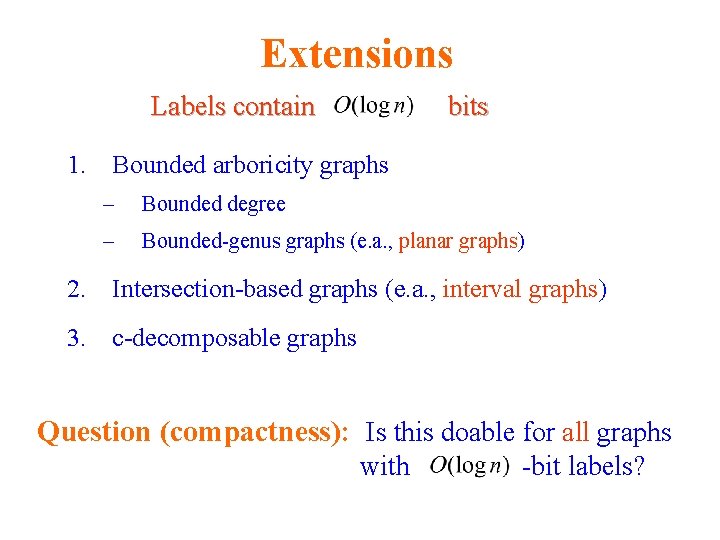
Extensions Labels contain bits 1. Bounded arboricity graphs – Bounded degree – Bounded-genus graphs (e. a. , planar graphs) 2. Intersection-based graphs (e. a. , interval graphs) 3. c-decomposable graphs Question (compactness): Is this doable for all graphs with -bit labels?
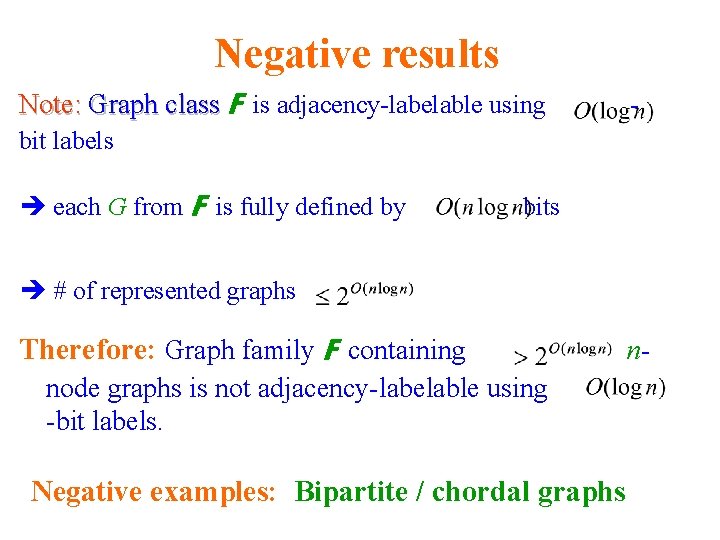
Negative results Note: Graph class F is adjacency-labelable using - bit labels each G from F is fully defined by bits # of represented graphs Therefore: Graph family F containing node graphs is not adjacency-labelable using -bit labels. Negative examples: Bipartite / chordal graphs n-

Question: Can this be done for other types of information beyond adjacency? • In general graphs • Distance • Edge / node connectivity • Routing • Flow • … • In trees • Ancestry • Lowest common ancestor (LCA or NCA) • Depth of NCA (separation level)

Question: Can this be done for other types of information beyond adjacency? • In general graphs • Distance • Edge / node connectivity • Routing • Flow • … • In trees • Ancestry • Lowest common ancestor (LCA or NCA) • Depth of NCA (separation level)
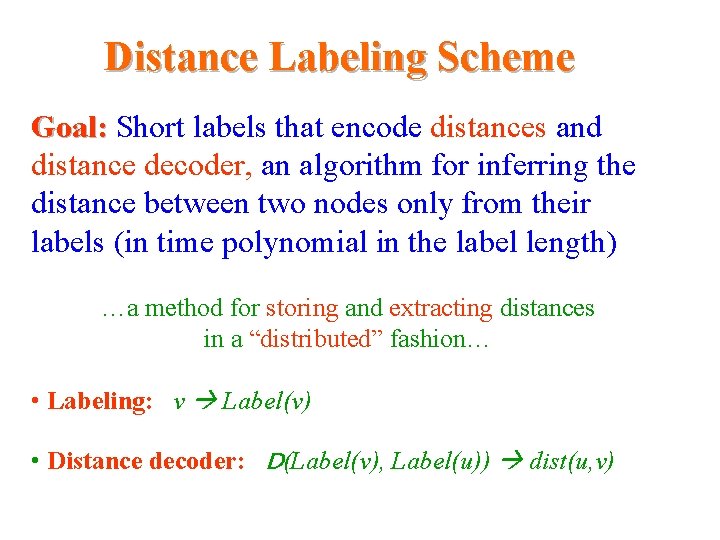
Distance Labeling Scheme Goal: Short labels that encode distances and distance decoder, an algorithm for inferring the distance between two nodes only from their Distance in the label length) labels (in time polynomial …a method for storing and extracting distances in a “distributed” fashion… • Labeling: v Label(v) • Distance decoder: D(Label(v), Label(u)) dist(u, v)
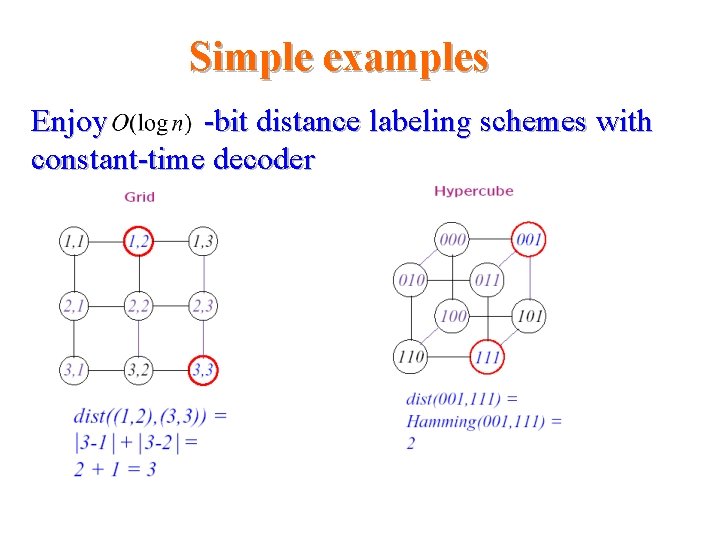
Simple examples Enjoy -bit distance labeling schemes with constant-time decoder Distance
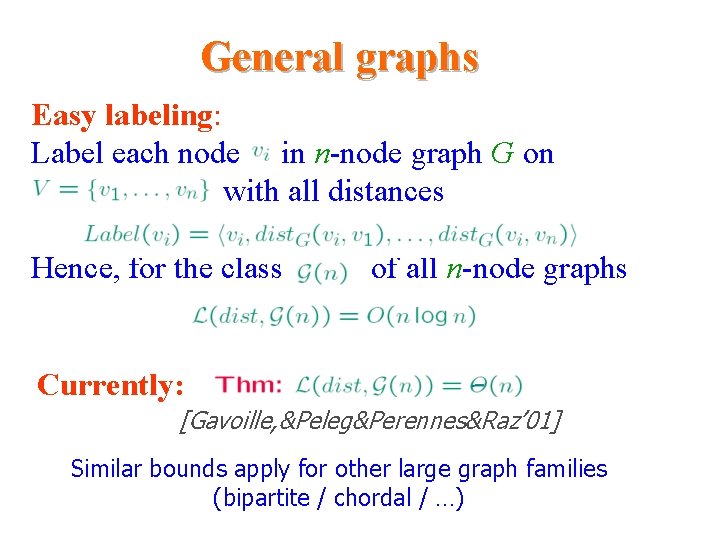
General graphs Easy labeling: Label each node in n-node graph G on with all distances Distance Hence, for the class of all n-node graphs Currently: [Gavoille, &Peleg&Perennes&Raz’ 01] Similar bounds apply for other large graph families (bipartite / chordal / …)
![Distance labeling on trees [Peleg’ 99] Preprocessing: Arbitrarily label each node v of T Distance labeling on trees [Peleg’ 99] Preprocessing: Arbitrarily label each node v of T](http://slidetodoc.com/presentation_image/1a50e3b647fe44c84dad3dc1d834cb50/image-16.jpg)
Distance labeling on trees [Peleg’ 99] Preprocessing: Arbitrarily label each node v of T with distinct integers I(v) from [1. . n]
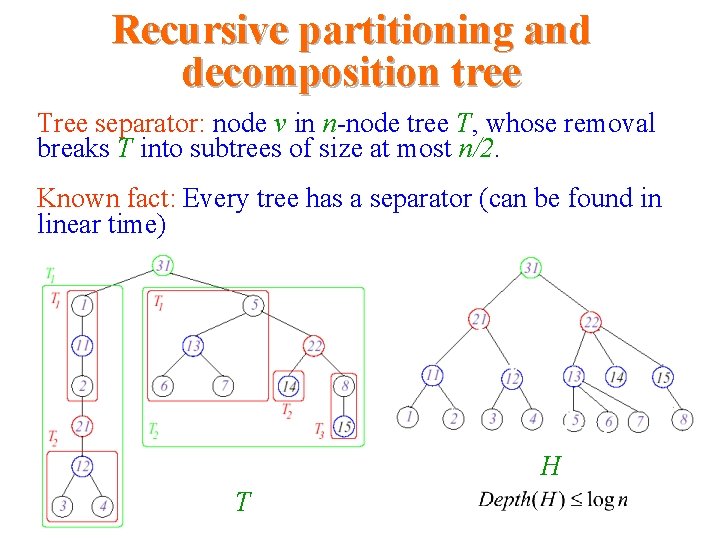
Recursive partitioning and decomposition tree Tree separator: node v in n-node tree T, whose removal breaks T into subtrees of size at most n/2. Known fact: Every tree has a separator (can be found in linear time) H T
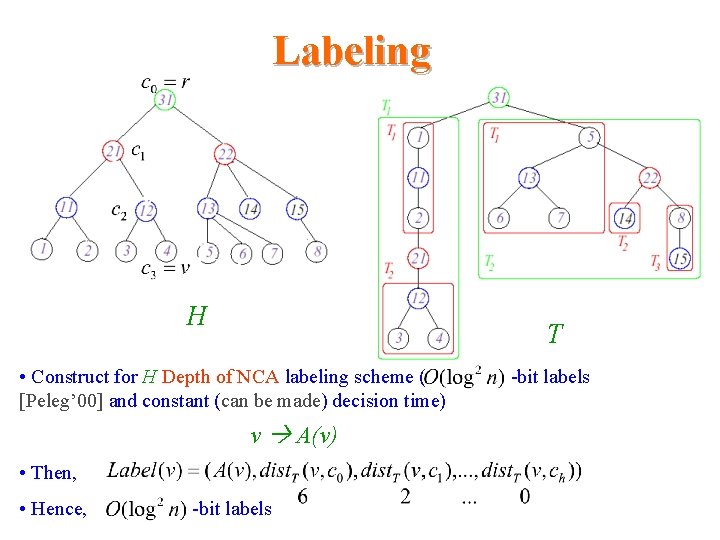
Labeling H T • Construct for H Depth of NCA labeling scheme ( [Peleg’ 00] and constant (can be made) decision time) v A(v) • Then, • Hence, -bit labels
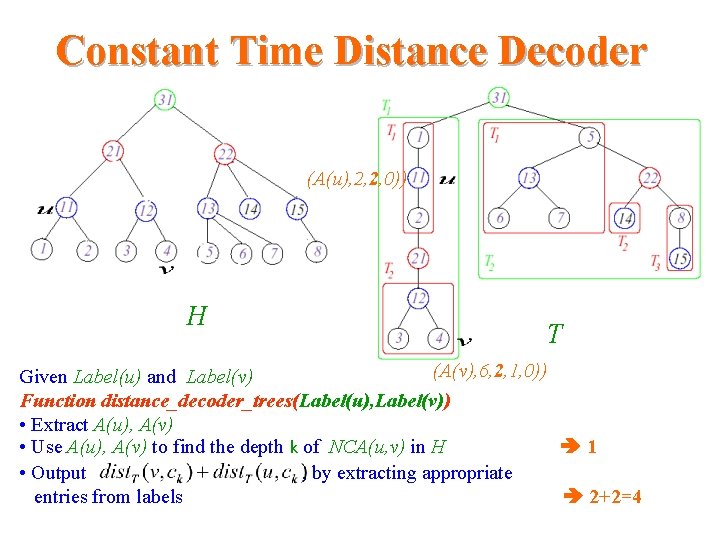
Constant Time Distance Decoder (A(u), 2, 2, 0)) H T (A(v), 6, 2, 1, 0)) Given Label(u) and Label(v) Function distance_decoder_trees(Label(u), Label(v)) • Extract A(u), A(v) • Use A(u), A(v) to find the depth k of NCA(u, v) in H 1 • Output , by extracting appropriate entries from labels 2+2=4
![Lower bounds and Planar graphs [Gavoille, &Peleg&Perennes&Raz’ 01] Lower bounds and Planar graphs [Gavoille, &Peleg&Perennes&Raz’ 01]](http://slidetodoc.com/presentation_image/1a50e3b647fe44c84dad3dc1d834cb50/image-20.jpg)
Lower bounds and Planar graphs [Gavoille, &Peleg&Perennes&Raz’ 01]
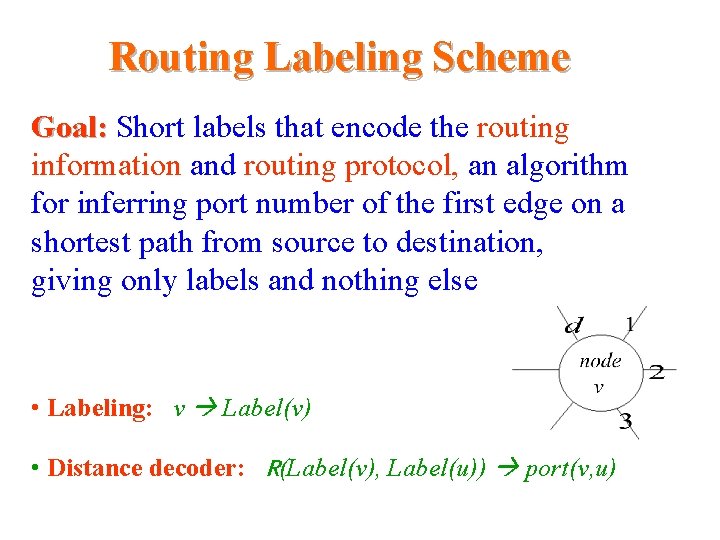
Routing Labeling Scheme Goal: Short labels that encode the routing information and routing protocol, an algorithm for inferring port number of the first edge on a shortest path from. Distance source to destination, giving only labels and nothing else • Labeling: v Label(v) • Distance decoder: R(Label(v), Label(u)) port(v, u)
General graphs Easy labeling (full routing tables): Label each node in n-node graph G on with full routing information Distance Hence, for the class of all n-node graphs one needs nlogn -bit labels Currently: Nothing better can be done for general graphs if we insist on routing via shortest paths
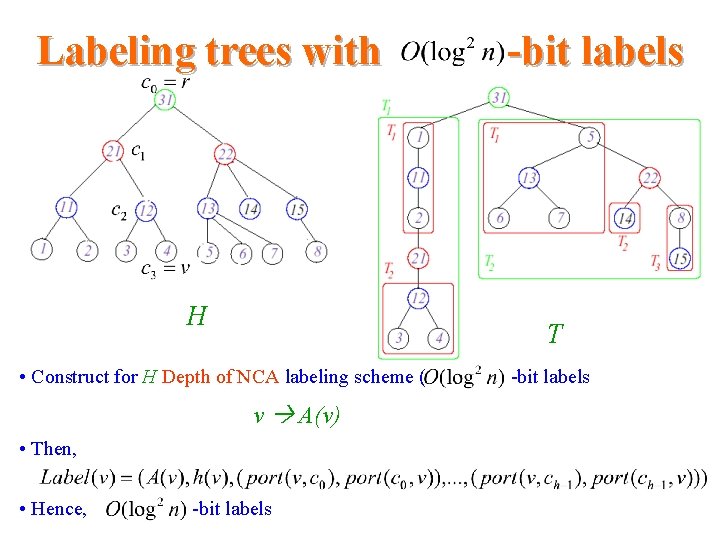
Labeling trees with H T • Construct for H Depth of NCA labeling scheme ( v A(v) • Then, • Hence, -bit labels
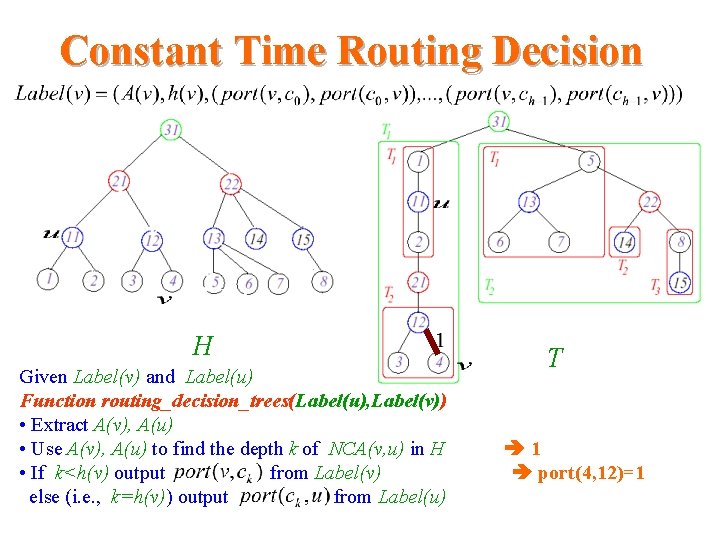
Constant Time Routing Decision H Given Label(v) and Label(u) Function routing_decision_trees(Label(u), Label(v)) • Extract A(v), A(u) • Use A(v), A(u) to find the depth k of NCA(v, u) in H • If k<h(v) output from Label(v) else (i. e. , k=h(v)) output from Label(u) T 1 port(4, 12)=1
![Best routing labeling scheme for trees and Planar graphs [Thorup&Zwick’ 01] For family of Best routing labeling scheme for trees and Planar graphs [Thorup&Zwick’ 01] For family of](http://slidetodoc.com/presentation_image/1a50e3b647fe44c84dad3dc1d834cb50/image-25.jpg)
Best routing labeling scheme for trees and Planar graphs [Thorup&Zwick’ 01] For family of n-node trees there is a routing labeling scheme with labels of size O(logn)-bits per node and constant time routing decision. [Fraigniaud&Gavoille’ 01] For family of n-node trees there is a routing labeling scheme with labels of size routing decision. -bits per node and constant time [Gavoille&Hanusse’ 99] For family of n-node planar graphs there is a routing labeling scheme with labels of size (8 n +o(n))-bits per node.
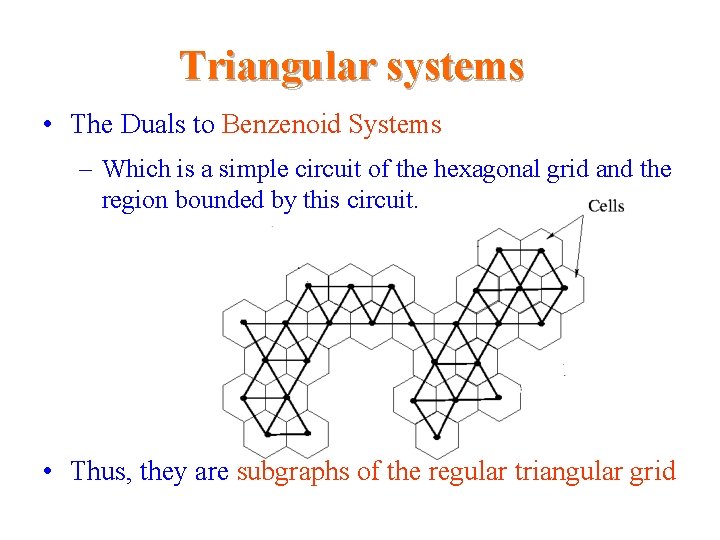
Triangular systems • The Duals to Benzenoid Systems – Which is a simple circuit of the hexagonal grid and the region bounded by this circuit. • Thus, they are subgraphs of the regular triangular grid
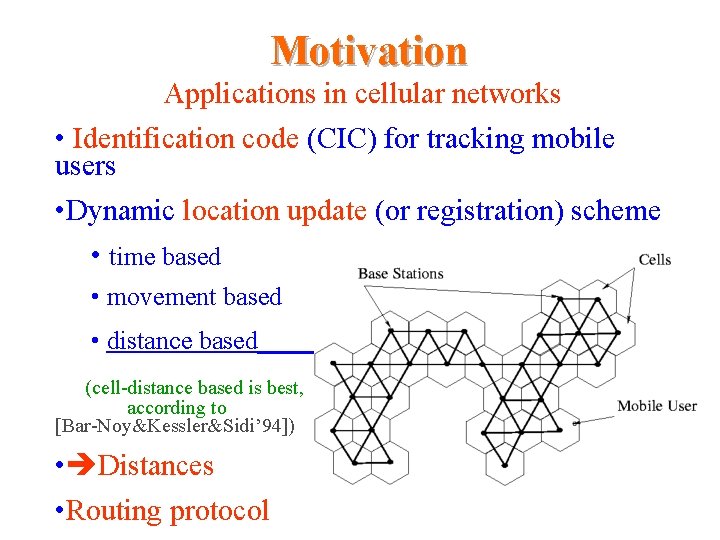
Motivation Applications in cellular networks • Identification code (CIC) for tracking mobile users • Dynamic location update (or registration) scheme • time based • movement based • distance based (cell-distance based is best, according to [Bar-Noy&Kessler&Sidi’ 94]) • Distances • Routing protocol
Current situation • Current cellular networks do not provide information that can be used to derive cell distances – It is hard to compute the distances between cells (claim from [Bar-Noy&Kessler&Sidi’ 94]) – It requires a lot of storage to maintain the distance information among cells (claim from [Akyildiz&Ho&Lin’ 96] and [Li&Kameda&Li’ 00])
![Recent results [Nocetti&Stojmenovic&Zhang’ 02] recently considered isometric subgraphs of the regular triangular grid and Recent results [Nocetti&Stojmenovic&Zhang’ 02] recently considered isometric subgraphs of the regular triangular grid and](http://slidetodoc.com/presentation_image/1a50e3b647fe44c84dad3dc1d834cb50/image-29.jpg)
Recent results [Nocetti&Stojmenovic&Zhang’ 02] recently considered isometric subgraphs of the regular triangular grid and give among others – A new cell addressing scheme using only three small integers, one of them being zero – A very simple method to compute the distance between two sells – A short and elegant routing protocol isometric not isometric
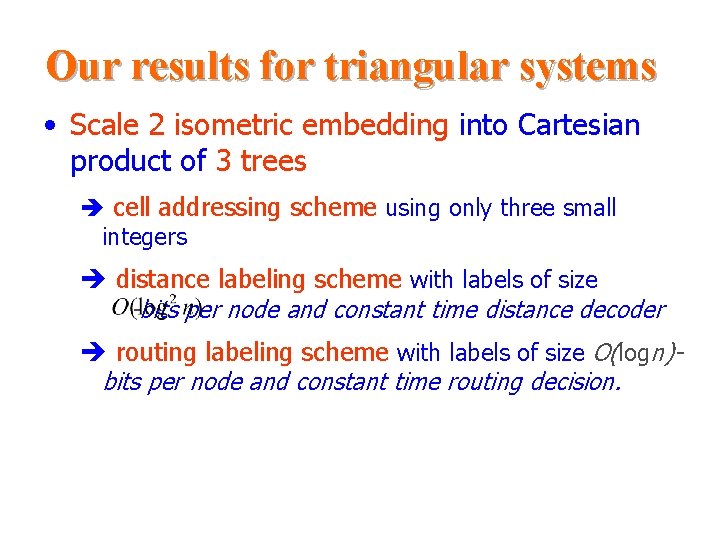
Our results for triangular systems • Scale 2 isometric embedding into Cartesian product of 3 trees cell addressing scheme using only three small integers distance labeling scheme with labels of size -bits per node and constant time distance decoder routing labeling scheme with labels of size O(logn)bits per node and constant time routing decision.
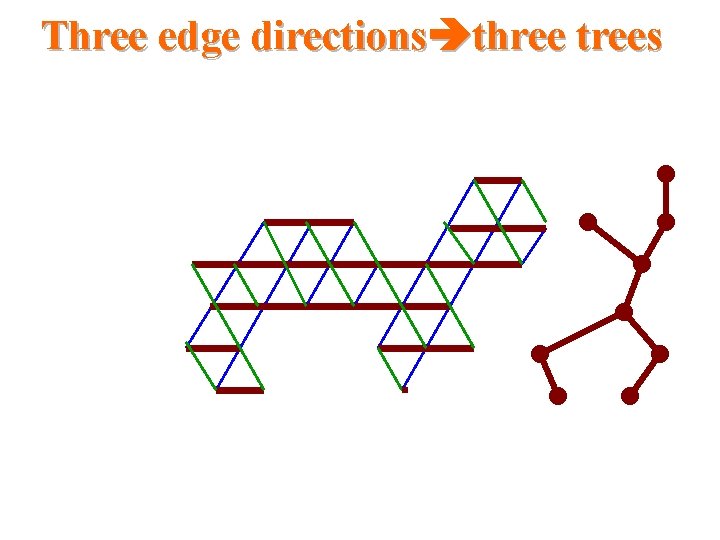
Three edge directions three trees
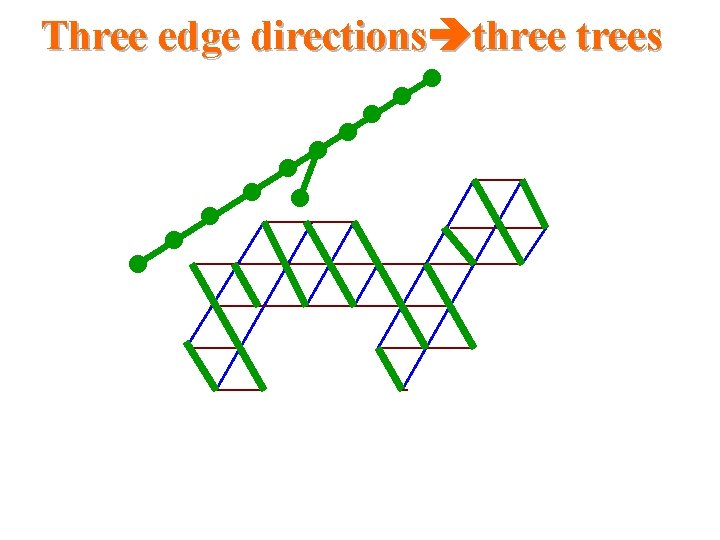
Three edge directions three trees
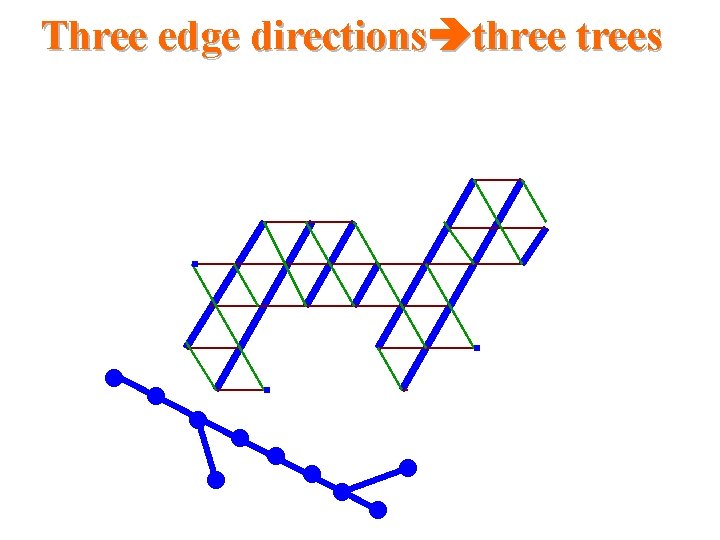
Three edge directions three trees
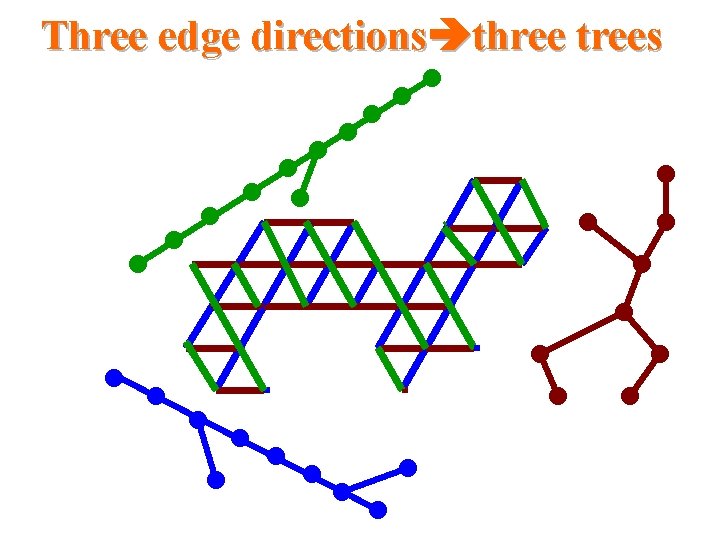
Three edge directions three trees
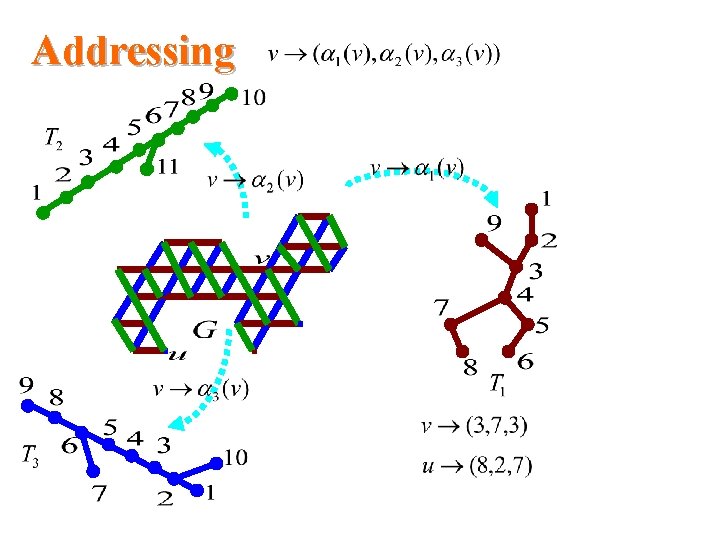
Addressing
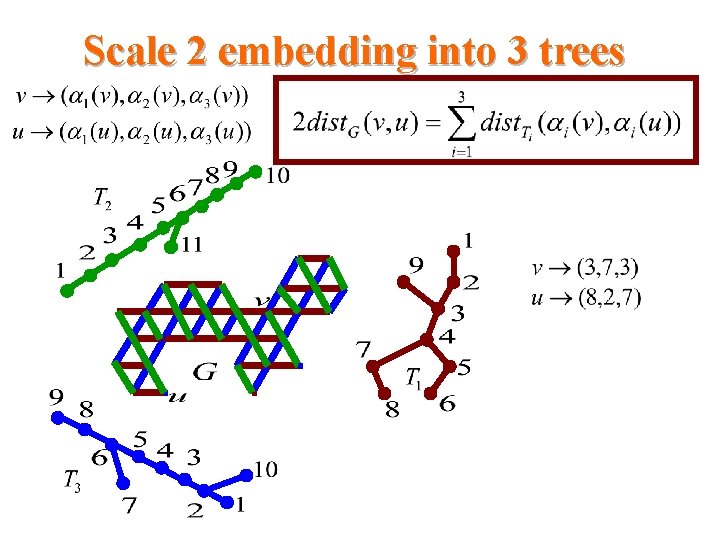
Scale 2 embedding into 3 trees
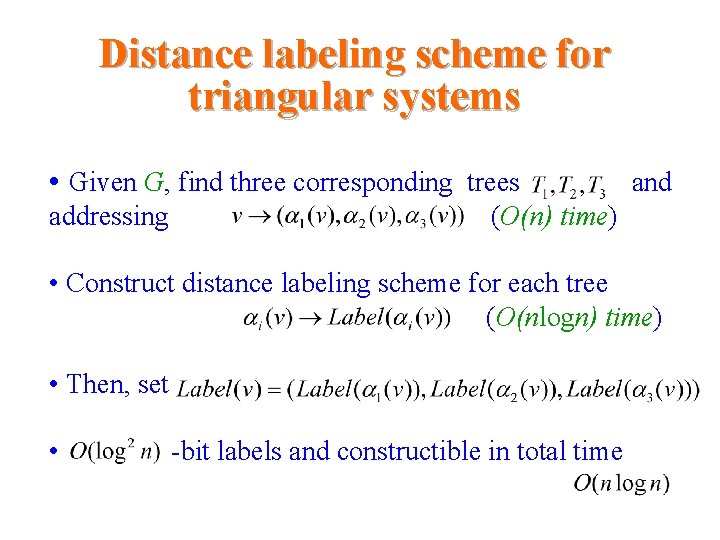
Distance labeling scheme for triangular systems • Given G, find three corresponding trees addressing and (O(n) time) • Construct distance labeling scheme for each tree (O(nlogn) time) • Then, set • -bit labels and constructible in total time
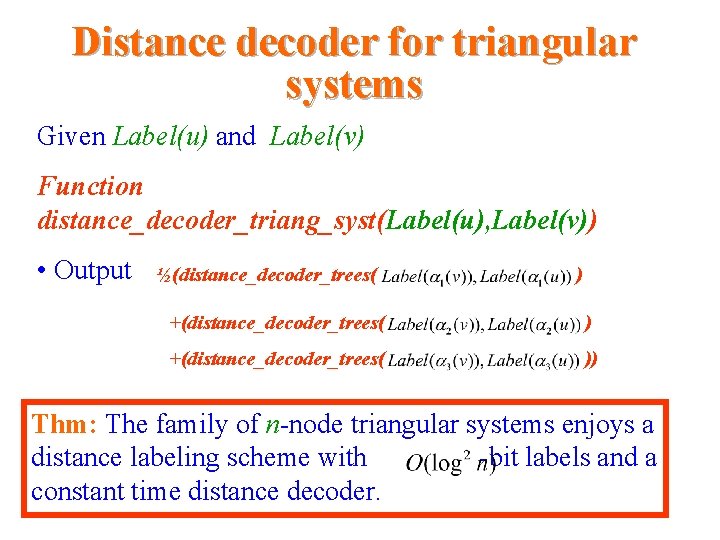
Distance decoder for triangular systems Given Label(u) and Label(v) Function distance_decoder_triang_syst(Label(u), Label(v)) • Output ½(distance_decoder_trees( ) +(distance_decoder_trees( )) Thm: The family of n-node triangular systems enjoys a distance labeling scheme with -bit labels and a constant time distance decoder.

Routing labeling scheme for triangular systems • Given G, find three corresponding trees and addressing • Construct routing labeling scheme for each tree using Thorup&Zwick method (log n bit labels) • Then, set Something more
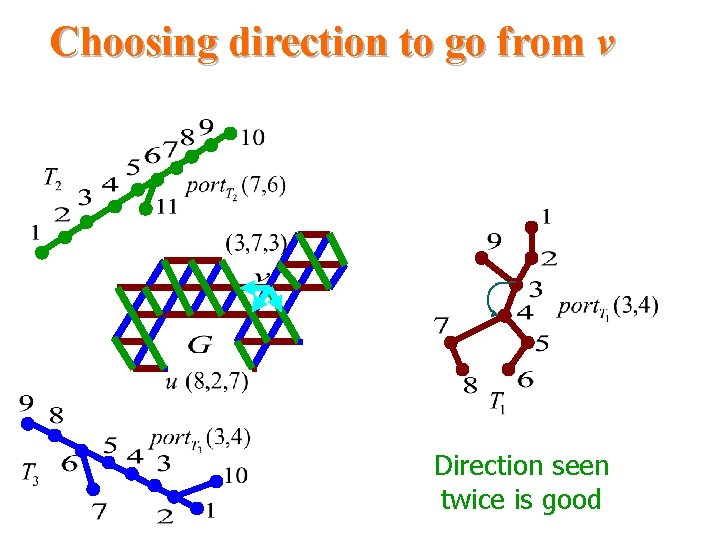
Choosing direction to go from v Direction seen twice is good
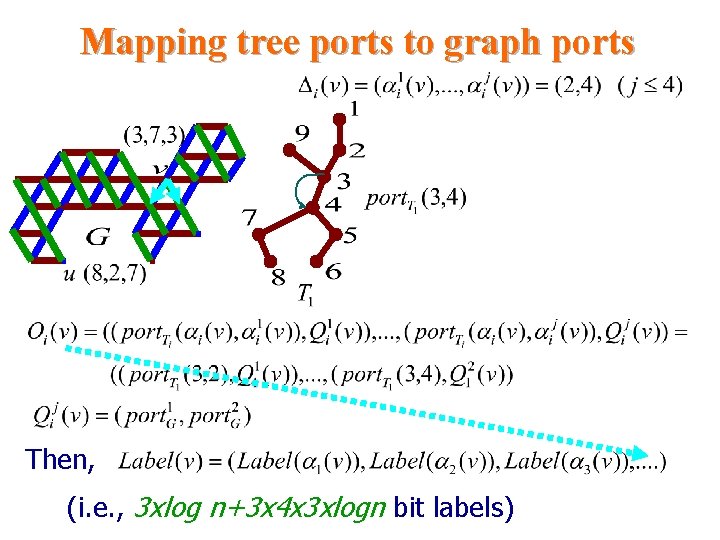
Mapping tree ports to graph ports Then, (i. e. , 3 xlog n+3 x 4 x 3 xlogn bit labels)
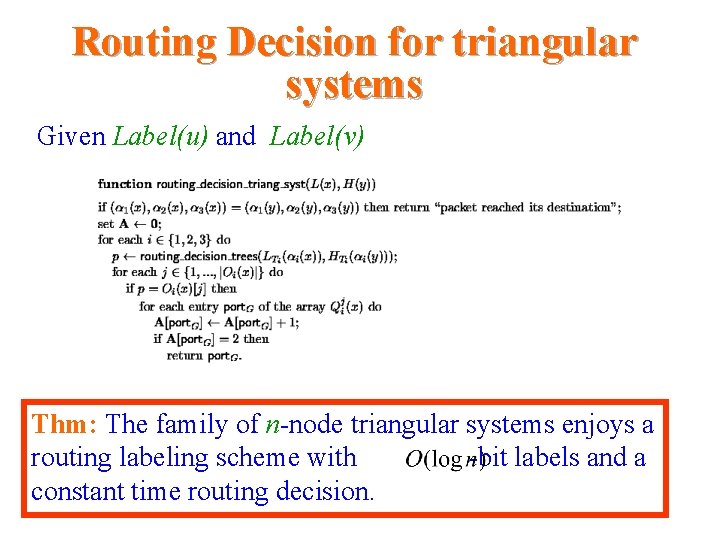
Routing Decision for triangular systems Given Label(u) and Label(v) Thm: The family of n-node triangular systems enjoys a routing labeling scheme with -bit labels and a constant time routing decision.
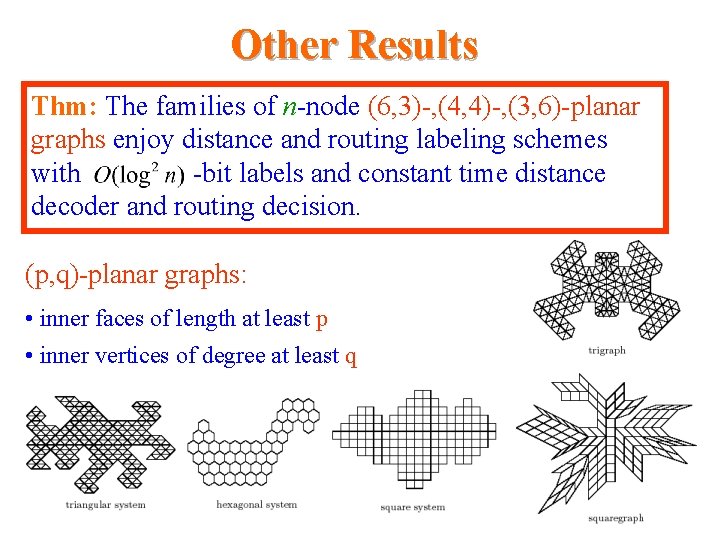
Other Results Thm: The families of n-node (6, 3)-, (4, 4)-, (3, 6)-planar graphs enjoy distance and routing labeling schemes with -bit labels and constant time distance decoder and routing decision. (p, q)-planar graphs: • inner faces of length at least p • inner vertices of degree at least q
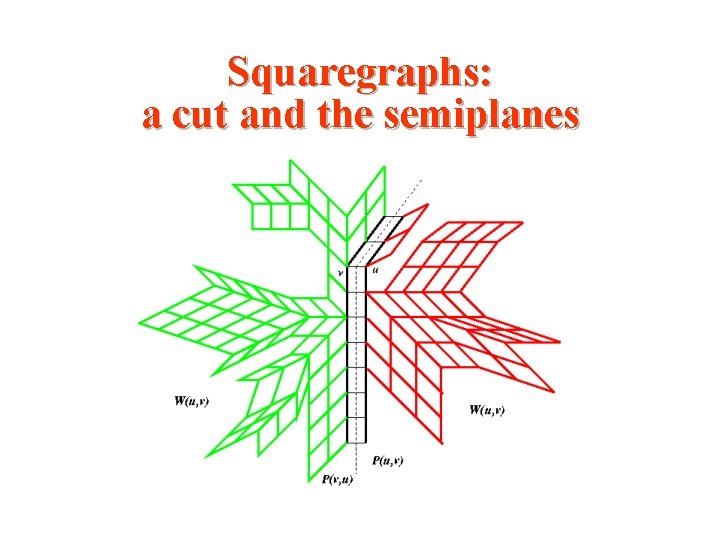
Squaregraphs: a cut and the semiplanes
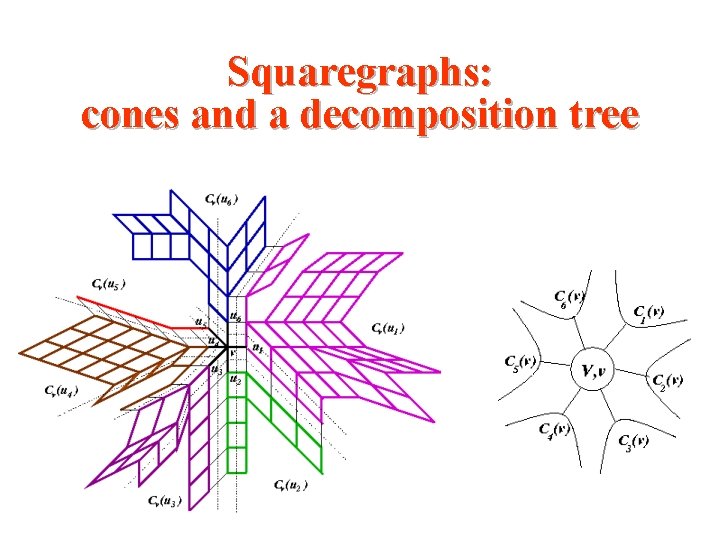
Squaregraphs: cones and a decomposition tree
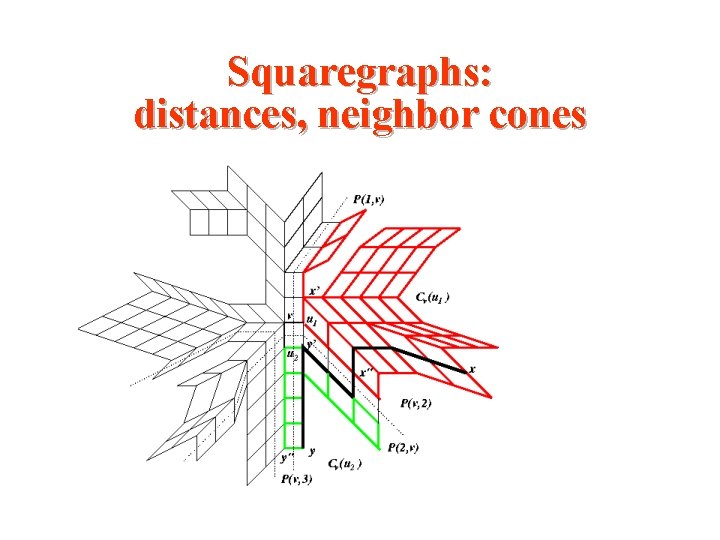
Squaregraphs: distances, neighbor cones

Thank you
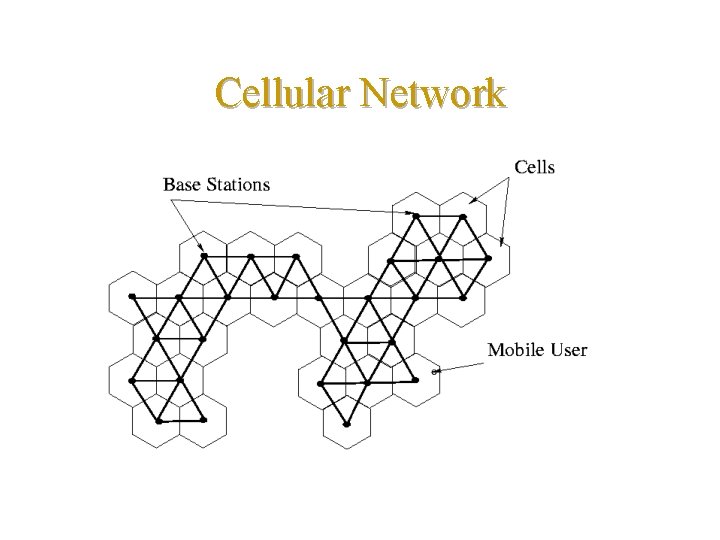
Cellular Network
- Slides: 48