Discovery of a putative causal mutation for Angus




- Slides: 27

Discovery of a putative causal mutation for Angus dwarfism Department of Animal Science NBCEC Brown Bagger Nov. 2006

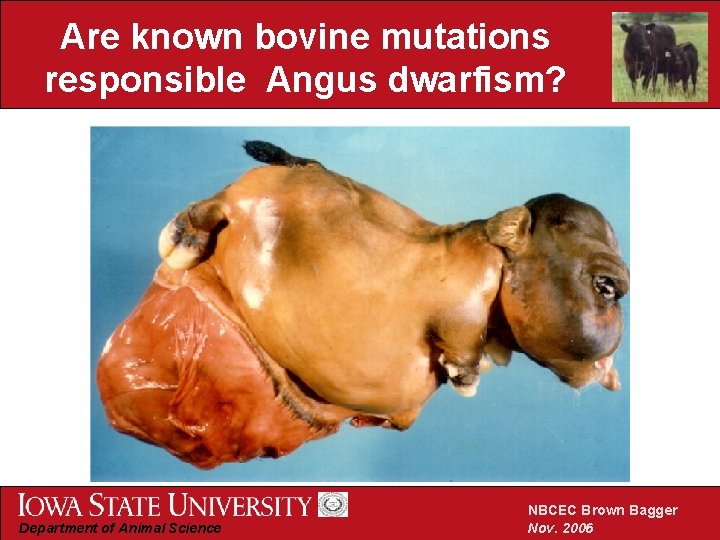
Summary of phenotypic characteristics • Shortened limbs, but normal body size • Decreased long bone and vertebral length • Incorrect endochondral ossification • Premature calcification of chondrocytes • Disorganized arrangement of osteocytes • Abnormal osteocyte phenotype Department of Animal Science NBCEC Brown Bagger Nov. 2006

Calves at 7 months vs. birth Two bulls at Birth 7 Months 16 c. M Dwarf T/T Dwarf male C/T Normal Department of Animal Science 18 c. M Normal T/T Dwarf female NBCEC Brown Bagger Nov. 2006

Strategy for finding the gene mutation for dwarfism • Consider known bovine dwarfisms • Select candidate bovine chromosomes for linkage analysis • Find a suitable population for linkage analysis Department of Animal Science NBCEC Brown Bagger Nov. 2006

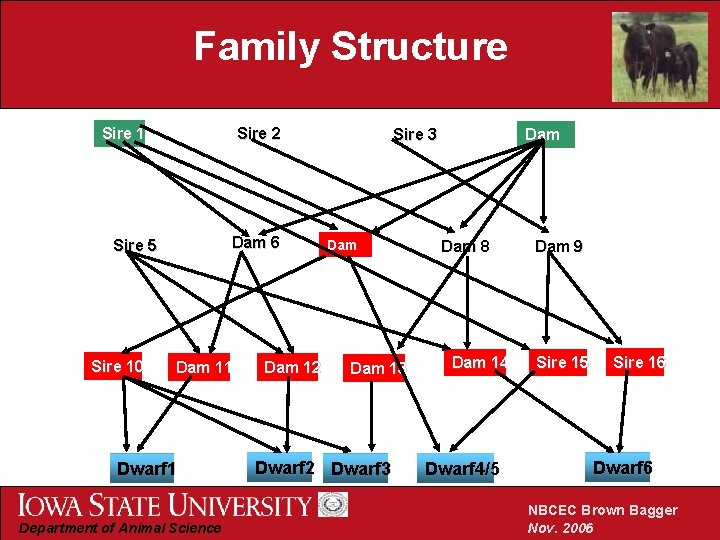
Family Structure Sire 1 Sire 2 Dam 6 Sire 5 Sire 10 Dam 11 Dwarf 1 Department of Animal Science Dam 12 Sire 3 Dam 7 Dam 13 Dwarf 2 Dwarf 3 Dam 4 Dam 8 Dam 14 Dwarf 4/5 Dam 9 Sire 15 Sire 16 Dwarf 6 NBCEC Brown Bagger Nov. 2006

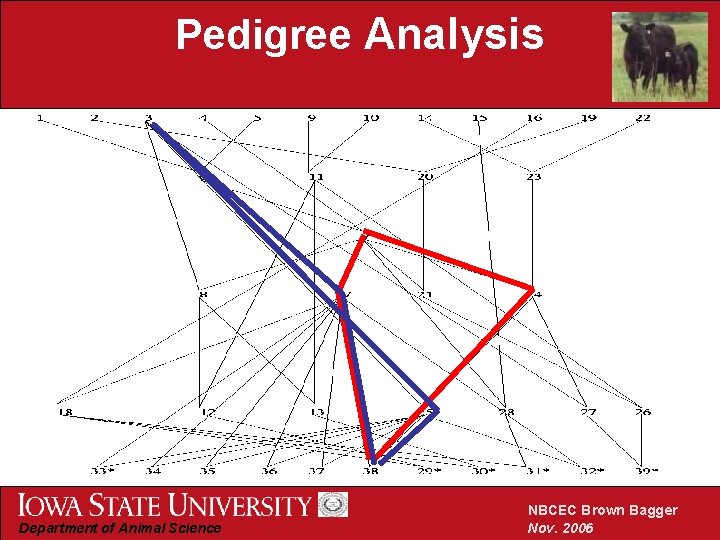
Pedigree Analysis Department of Animal Science NBCEC Brown Bagger Nov. 2006

Are known bovine mutations responsible Angus dwarfism? 1356 TTTTCAAAGG C GAGCTGAAGC - Normal limbin TTTTCAAAGG T GAGCTGAAGC - Mutant limbin TTTTCAAAGG C GAGCTGAAGC - Angus dwarf/normal 2054/2055 AGGGACCTGG CA GCCAGGGCCG - Normal limbin AGGGACCTGG G GCCAGGGCCG - Mutant limbin AGGGACCTGG CA GCCAGGGCCG – Angus dwarf/normal Mishra et al. , 2003 Department of Animal Science NBCEC Brown Bagger Nov. 2006

Are known bovine mutations responsible Angus dwarfism? Department of Animal Science NBCEC Brown Bagger Nov. 2006

Whole Genome Scan • Systematically evaluate the bovine genome • Chromosomes known to contain genes implicated in dwarfism in other species were given a priority for evaluation • Evenly spaced microsatellites – Genotyping done in Dr. Georges lab in Belgium Department of Animal Science NBCEC Brown Bagger Nov. 2006

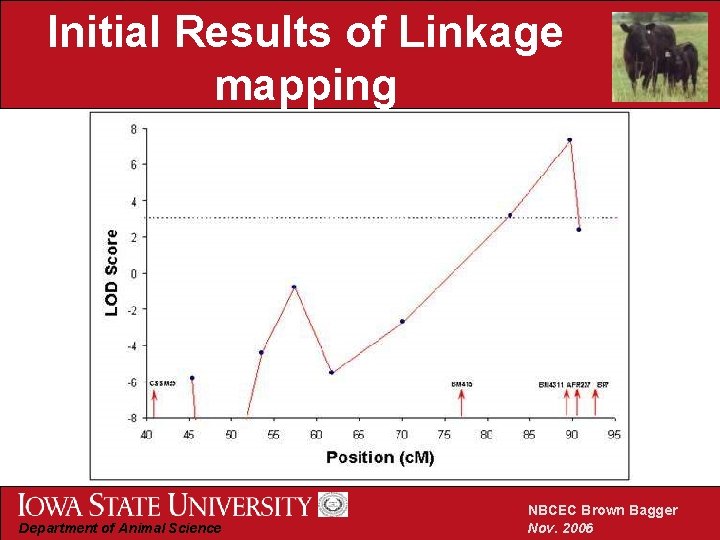
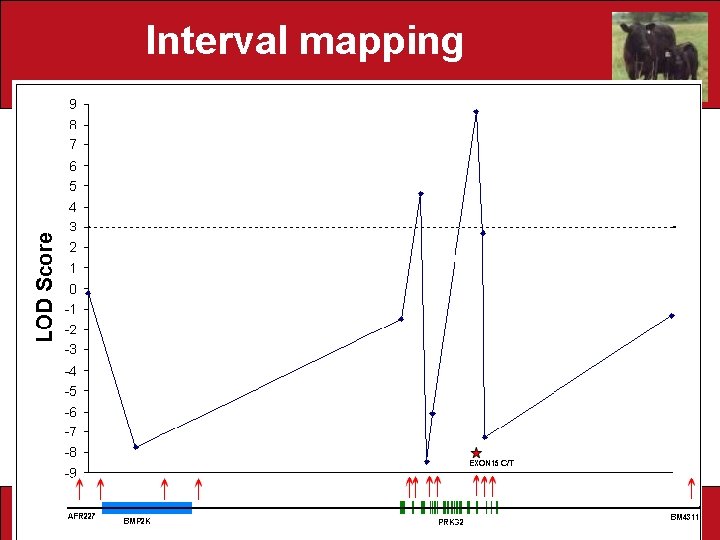
Pinpointing the genomic region associated with dwarfism • Tested marker intervals for Likelihood of odds (LOD) with dwarfism LOD =Log 10 Marker Interval contains dwarf mutation Dwarf mutation is on a different chromosome • Critical value: LOD=3. 0 • Problem: missing genotypes Department of Animal Science NBCEC Brown Bagger Nov. 2006

Initial Results of Linkage mapping Department of Animal Science NBCEC Brown Bagger Nov. 2006

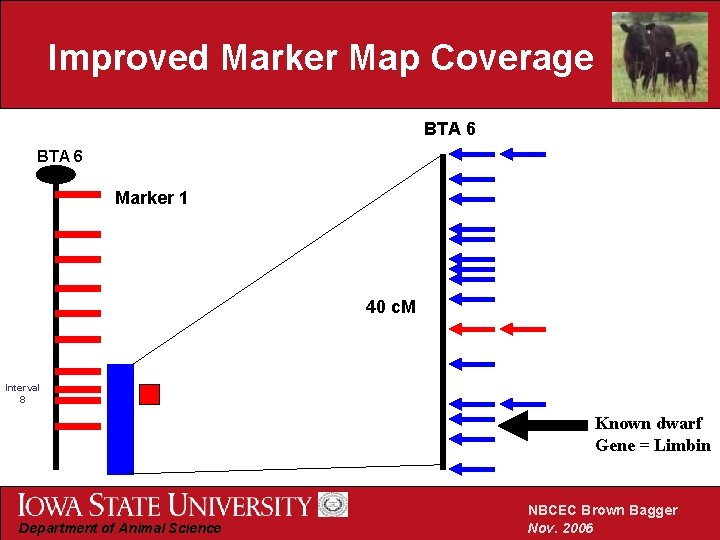
Improved Marker Map Coverage BTA 6 Marker 1 40 c. M Interval 8 Known dwarf Gene = Limbin Department of Animal Science NBCEC Brown Bagger Nov. 2006

Summary of initial results • Known dwarf mutations are not responsible for Angus dwarfism • Linkage detected to BTA 6 • Limbin eliminated as a candidate gene • Critical region refined to 2. 8 c. M Department of Animal Science NBCEC Brown Bagger Nov. 2006

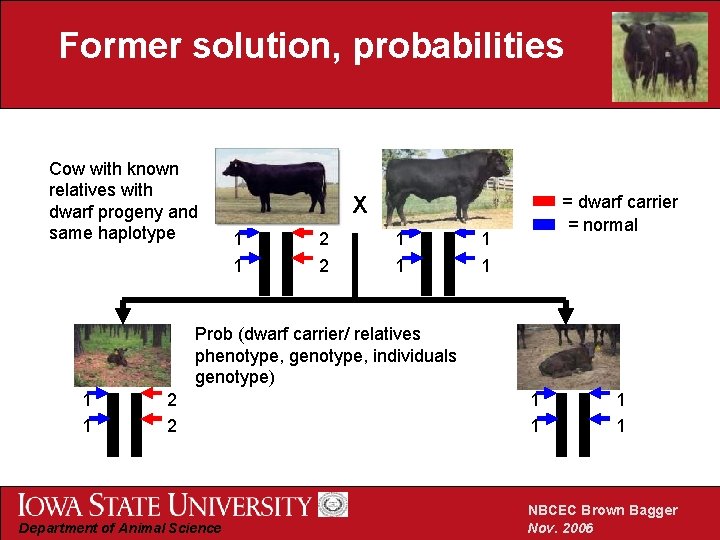
Former solution, probabilities Cow with known relatives with dwarf progeny and same haplotype = dwarf carrier = normal X 1 1 2 2 1 1 Prob (dwarf carrier/ relatives phenotype, genotype, individuals genotype) 1 1 2 2 Department of Animal Science 1 1 NBCEC Brown Bagger Nov. 2006

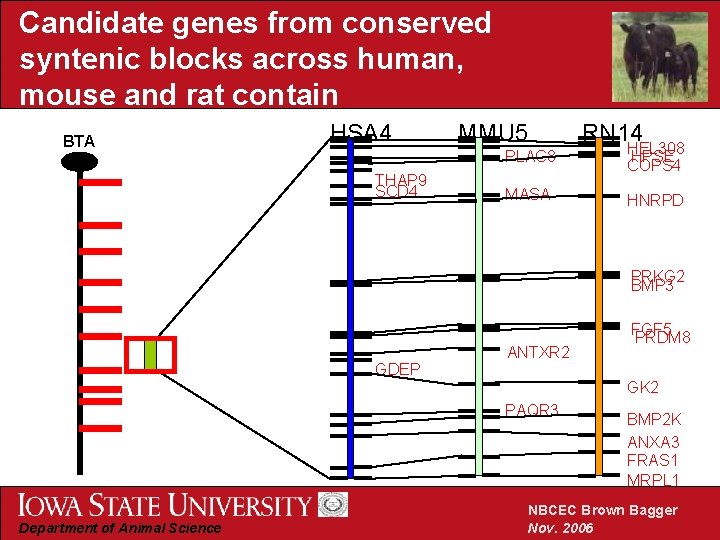
Candidate genes from conserved syntenic blocks across human, mouse and rat contain BTA HSA 4 THAP 9 SCD 4 MMU 5 RN 14 PLAC 8 HEL 308 HPSE COPS 4 MASA HNRPD PRKG 2 BMP 3 GDEP ANTXR 2 GK 2 PAQR 3 Department of Animal Science FGF 5 PRDM 8 BMP 2 K ANXA 3 FRAS 1 MRPL 1 NBCEC Brown Bagger Nov. 2006

Selection of candidate genes • • Gene Function Knockout analysis in the mouse Naturally occurring mutants Based on these criteria, 4 genes were selected Department of Animal Science NBCEC Brown Bagger Nov. 2006

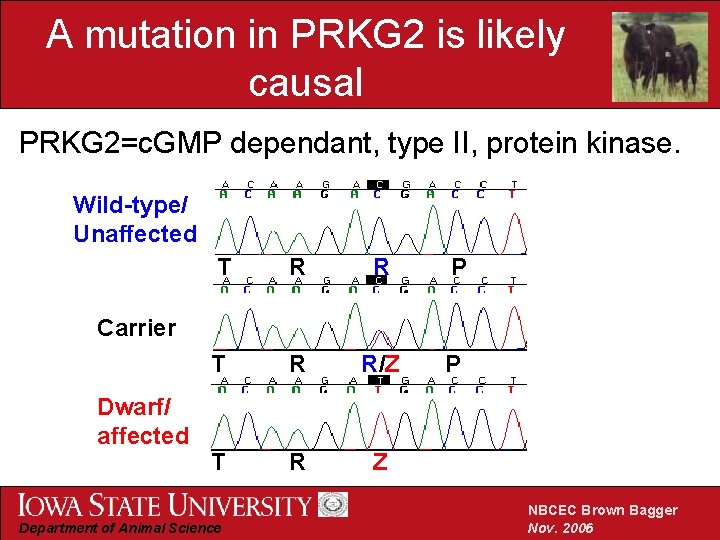
Overview of positional candidate genes • Bone Morphogenetic Protein 2 Kinase (BMP 2 K) • Fibroblast Growth Factor 5 (FGF 5) • Bone Morphogenetic Protein 3 • c. GMP-dependant, type II, protein kinase (PRKG 2) Department of Animal Science NBCEC Brown Bagger Nov. 2006

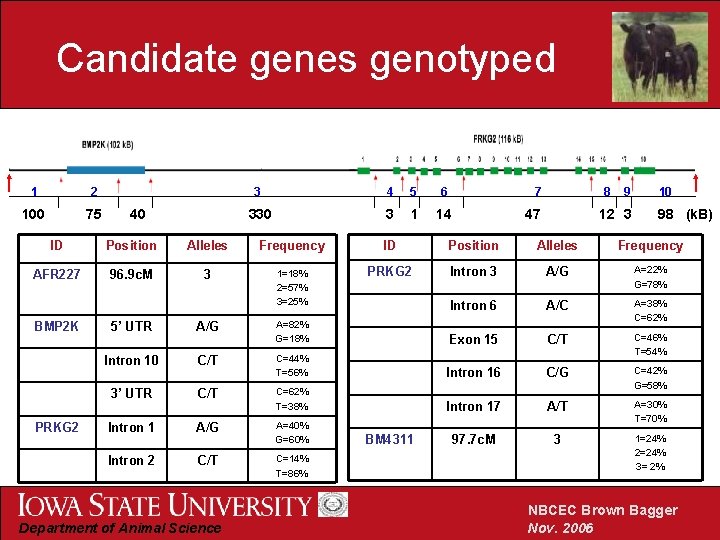
Candidate genes genotyped 1 2 100 75 40 3 4 5 6 7 330 3 1 14 8 47 9 12 3 10 98 (k. B) ID Position Alleles Frequency AFR 227 96. 9 c. M 3 1=18% 2=57% 3=25% PRKG 2 Intron 3 A/G A=22% G=78% Intron 6 A/C A=38% C=62% BMP 2 K PRKG 2 5’ UTR A/G A=82% G=18% Exon 15 C/T Intron 10 C/T C=44% T=56% C=46% T=54% Intron 16 C/G 3’ UTR C/T C=62% T=38% C=42% G=58% Intron 17 A/T Intron 1 A/G A=40% G=60% A=30% T=70% 97. 7 c. M 3 Intron 2 C/T C=14% T=86% 1=24% 2=24% 3= 2% Department of Animal Science BM 4311 NBCEC Brown Bagger Nov. 2006

Interval mapping Marker Interval Department of Animal Science NBCEC Brown Bagger Nov. 2006

A mutation in PRKG 2 is likely causal PRKG 2=c. GMP dependant, type II, protein kinase. Wild-type/ Unaffected T R R P T R R/Z P T R Z Carrier Dwarf/ affected Department of Animal Science NBCEC Brown Bagger Nov. 2006

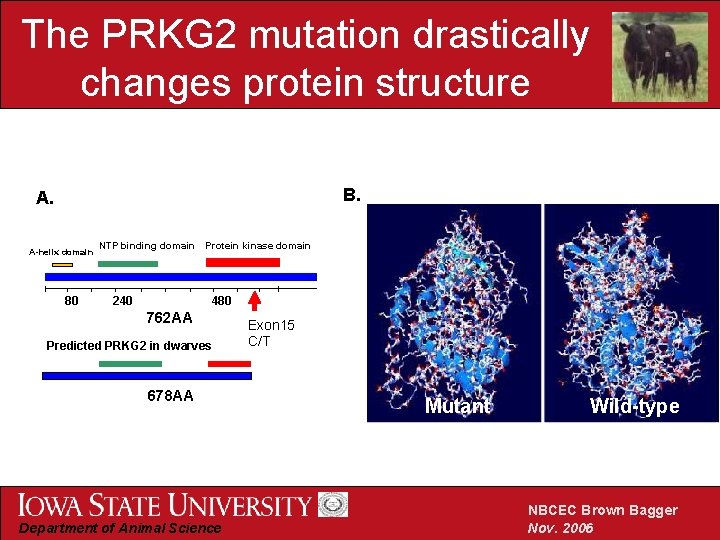
The PRKG 2 mutation drastically changes protein structure B. A. Α-helix domain 80 NTP binding domain Protein kinase domain 240 480 762 AA Predicted PRKG 2 in dwarves 678 AA Department of Animal Science Exon 15 C/T Mutant Wild-type NBCEC Brown Bagger Nov. 2006

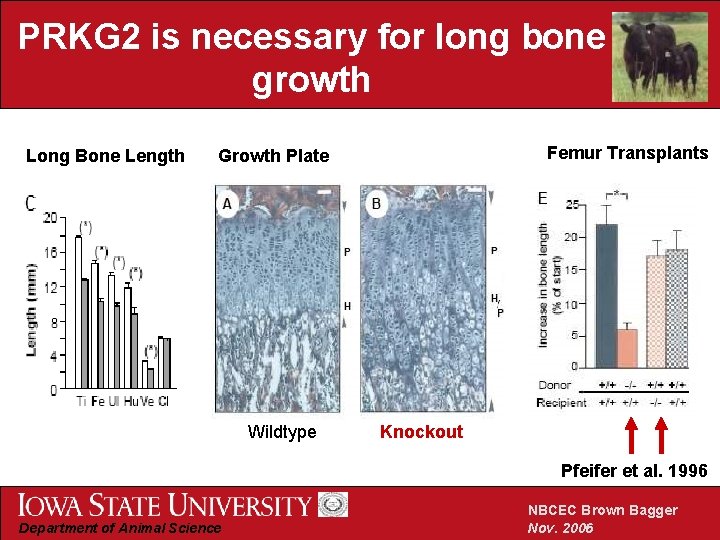
PRKG 2 is necessary for long bone growth Long Bone Length Femur Transplants Growth Plate Wildtype Knockout Pfeifer et al. 1996 Department of Animal Science NBCEC Brown Bagger Nov. 2006

Additional functional work Female dwarf Department of Animal Science Male dwarf Male carrier NBCEC Brown Bagger Nov. 2006

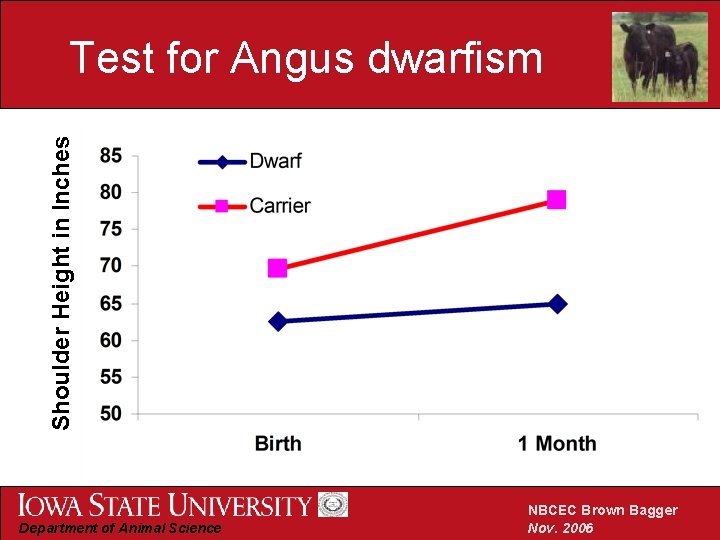
Shoulder Height in Inches Test for Angus dwarfism Department of Animal Science NBCEC Brown Bagger Nov. 2006

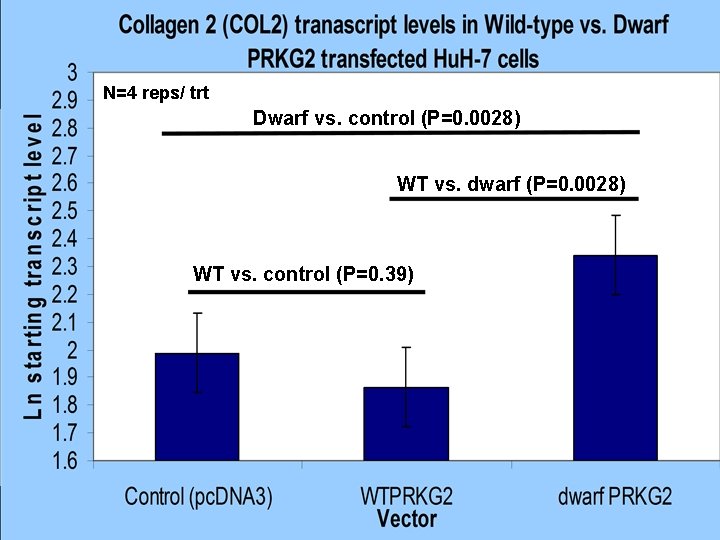
N=4 reps/ trt Dwarf vs. control (P=0. 0028) WT vs. dwarf (P=0. 0028) WT vs. control (P=0. 39) Department of Animal Science NBCEC Brown Bagger Nov. 2006

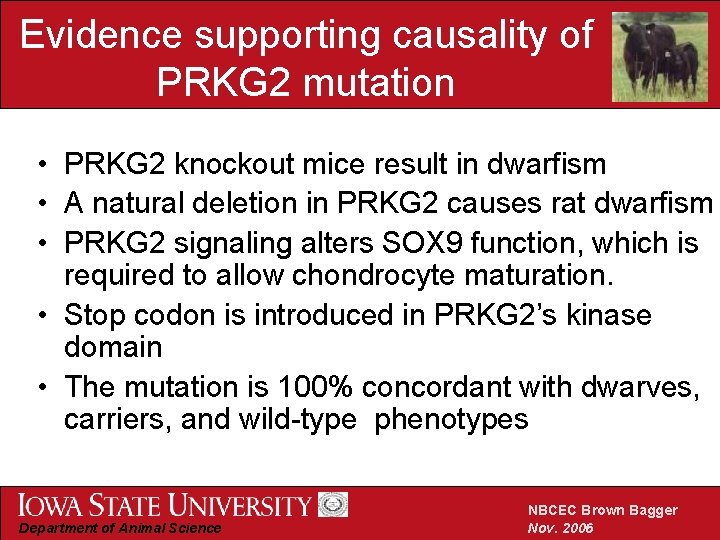
Evidence supporting causality of PRKG 2 mutation • PRKG 2 knockout mice result in dwarfism • A natural deletion in PRKG 2 causes rat dwarfism • PRKG 2 signaling alters SOX 9 function, which is required to allow chondrocyte maturation. • Stop codon is introduced in PRKG 2’s kinase domain • The mutation is 100% concordant with dwarves, carriers, and wild-type phenotypes Department of Animal Science NBCEC Brown Bagger Nov. 2006

Acknowledgements Iowa State James Koltes Bishnu Mishra Dr. Zhiliang Hu University of Nebraska University of Liege University of Sydney Michel Georges Wouter Coppieters Julie Cavanagh Imke Tammen Haruko Takeda Ohio State University David Steffen Washington State University Chuck Hines Rowland Cobbold American Angus Association Tawfik Aboellail Department of Animal Science Bryce Schumann NBCEC Brown Bagger Nov. 2006