Discovering functional interaction patterns in ProteinProtein Interactions Networks



- Slides: 29

Discovering functional interaction patterns in Protein-Protein Interactions Networks ¡ ¡ Authors: Mehmet E Turnalp Tolga Can Presented By: Sandeep Kumar

Background ¡ Availability of genome scale protein network ¡ Understanding topological organization ¡ Identification of conserved subnetworks across different species ¡ Discover modules of interaction ¡ Predict functions of uncharacterized proteins ¡ Improve the accuracy of currently available networks

Aim of study ¡ Using available functional annotations of proteins in PPI network and look for overrepresented patterns of interactions in the network ¡ Present new frequent pattern identification technique PPISpan

Yeast as a model ¡ Why yeast genomics? A model eukaryote organism … ¡ Well known PPI network Saccharomyces cerevisiae

PPI Network ¡ Protein protein interaction shown by edge between them indicating physical association in the form of modification, transport or complex formation ¡ Interesting conserved interaction patterns among species ¡ Patterns correspond to specific biological process

Frequent sub-graphs A graph (sub graph) is frequent if is support (occurrence frequency) in a given dataset is no less than minimum support threshold

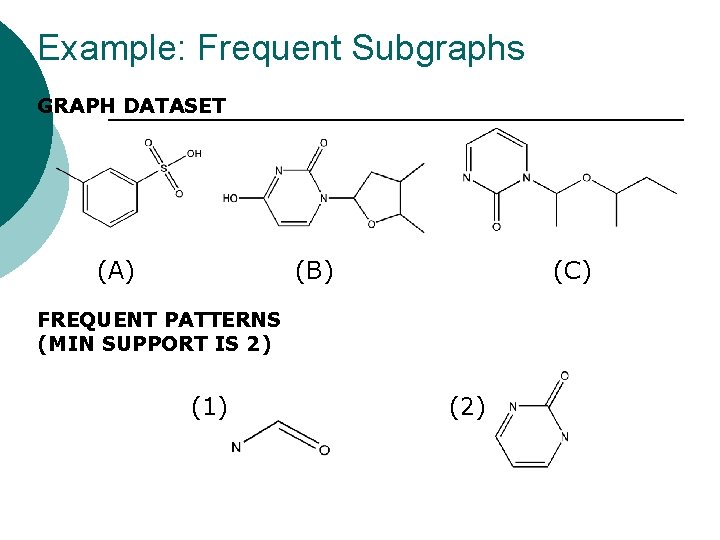
Example: Frequent Subgraphs GRAPH DATASET (A) (B) (C) FREQUENT PATTERNS (MIN SUPPORT IS 2) (1) (2)

The Algorithm - PPISpan ¡ Based on g. Span ¡ Modified to adapt for PPI network ¡ Candidate generation ¡ Frequency counting

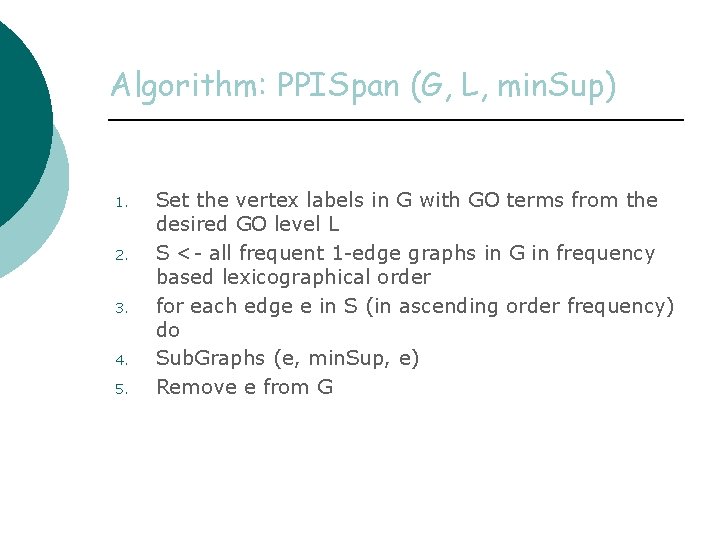
Algorithm: PPISpan (G, L, min. Sup) 1. 2. 3. 4. 5. Set the vertex labels in G with GO terms from the desired GO level L S <- all frequent 1 -edge graphs in G in frequency based lexicographical order for each edge e in S (in ascending order frequency) do Sub. Graphs (e, min. Sup, e) Remove e from G

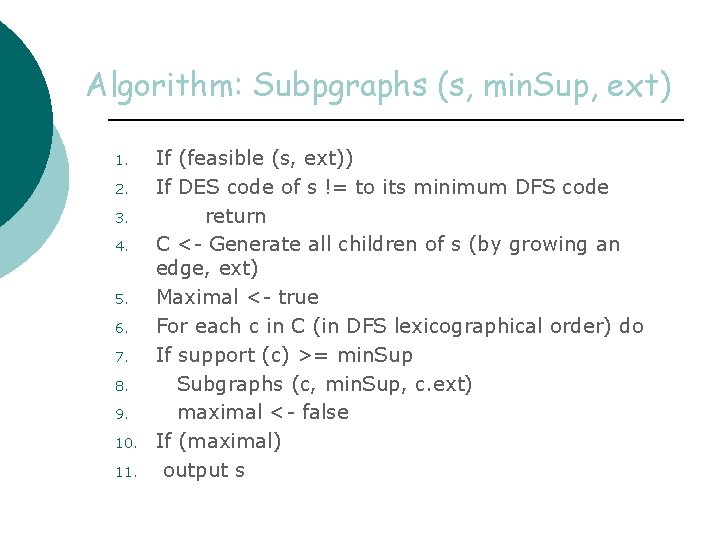
Algorithm: Subpgraphs (s, min. Sup, ext) 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. If (feasible (s, ext)) If DES code of s != to its minimum DFS code return C <- Generate all children of s (by growing an edge, ext) Maximal <- true For each c in C (in DFS lexicographical order) do If support (c) >= min. Sup Subgraphs (c, min. Sup, c. ext) maximal <- false If (maximal) output s

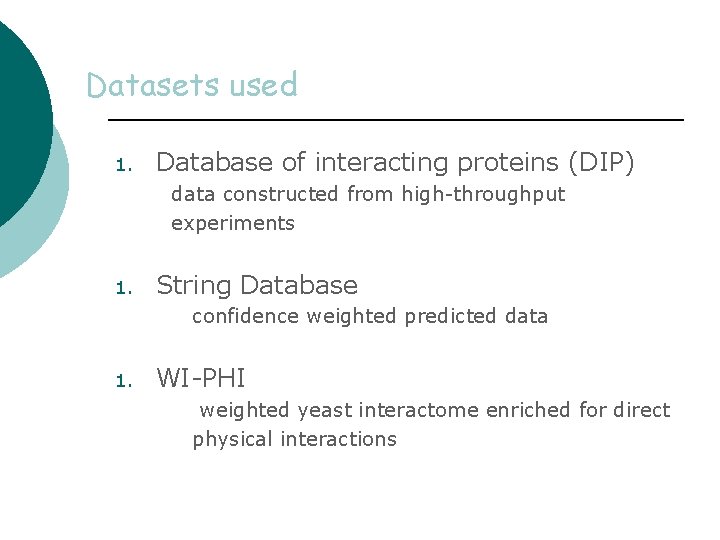
Datasets used 1. Database of interacting proteins (DIP) data constructed from high-throughput experiments 1. String Database confidence weighted predicted data 1. WI-PHI weighted yeast interactome enriched for direct physical interactions

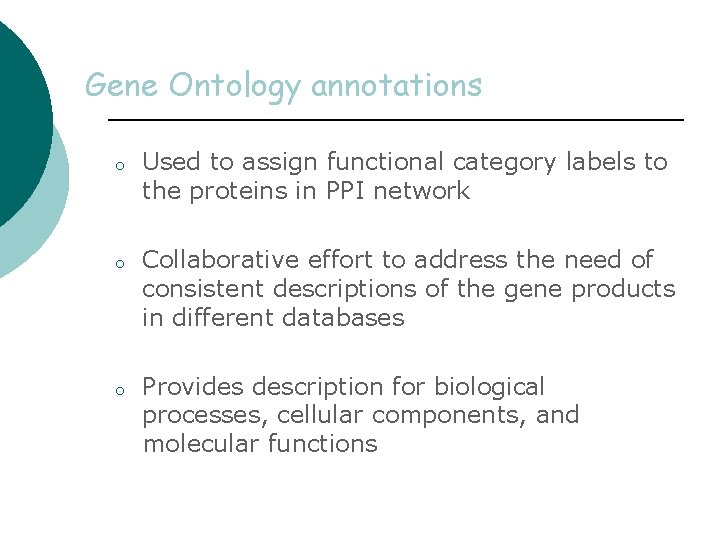
Gene Ontology annotations o o o Used to assign functional category labels to the proteins in PPI network Collaborative effort to address the need of consistent descriptions of the gene products in different databases Provides description for biological processes, cellular components, and molecular functions

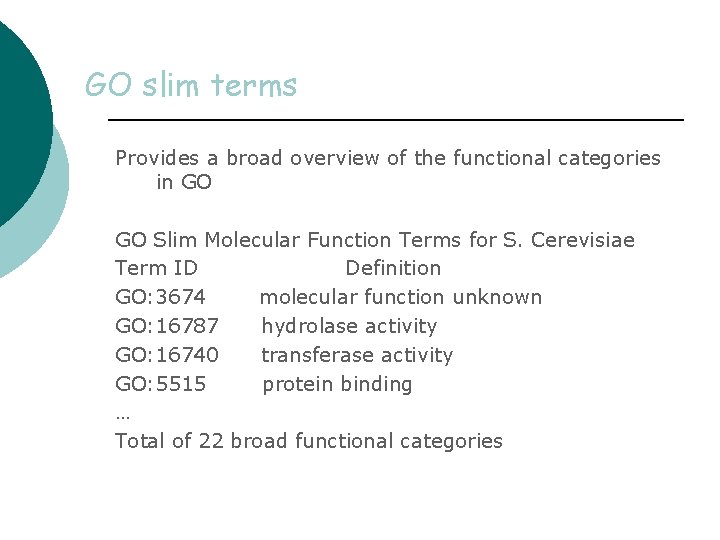
GO slim terms Provides a broad overview of the functional categories in GO GO Slim Molecular Function Terms for S. Cerevisiae Term ID Definition GO: 3674 molecular function unknown GO: 16787 hydrolase activity GO: 16740 transferase activity GO: 5515 protein binding … Total of 22 broad functional categories

Research Steps o o Label the nodes with functional categories with GO annotations Consider molecular function hierarchy Focus on functional interaction patterns in arbitrarily topologies Find non-overlapping embeddings using PPISpan

Problems faced o o Noise in PPI network False positives False negatives Accuracy and specificity of annotations of proteins

Supporting embedding o Specific instance of the functional pattern realized by certain proteins in the PPI network

Experiment details o o Implemented in C++ Searched for frequent interaction patterns of support >= 15

Pattern frequency in different datasets Number of patterns found

Observation ¡ Most of the patterns are trees ¡ Star topology most abundant ¡ Cycles rare

Comparison with known molecular complexes and pathways Ignore topology and treat patterns as set of proteins for comparison ¡ Molecular complexes from MIPS (Munich Information Center for Protein Sequences) complex catalogue database ¡ Signaling, transport, and regulatory pathways from KEGG database ¡ Use high quality complexes ¡

cpcount o o Average number of different complexes or pathways the embeddings of a frequent interaction pattern overlaps with To speculate on the location of interacting patterns

cpoverlap o o Quantifies the overlap between proteins in an embedding and known complexes and pathways Ratio of proteins in an embedding that are members of known functional modules

Observations from comparison o o o For some of the observed patterns, topology is more important than underlying functional annotations Comparison of all the patterns with random patterns in terms of overlap with MIPS complexes Comparison of all the patterns with random patterns in terms of overlap with transport and signaling pathways

Analysis of patterns with MIPS complexes o o Selected patterns from DIP and WI-PHI networks Selected patterns from the STRING network cpoverlap of selected patterns with respect to MIPS complexes cpcount of selected patterns with respect to MIPS complexes

Analysis of patterns with KEGG pathways o o o Selected patterns from DIP, STRING and WI -PHI networks cpoverlap of selected patterns with respect to transport and signaling pathways cpcount of selected patterns with respect to transport and signaling pathways

Some interesting Functional interaction patterns o o A frequent functional interaction pattern in the DIP network A frequent functional interaction pattern in the WI-PHI network A functional interaction pattern related to the MAPK signaling pathways A functional interaction pattern related to the SNARE interactions in vesicular transport

Conclusions o o Proposed new frequent pattern identification technique, PPISpan utilized molecular function Gene Ontology annotations to assign non-unique labels to proteins of a PPI network identified significantly frequent functional interaction patterns Frequent patterns offer a new perspective into the modular organization of protein interaction networks

QUESTIONS ?

THANK YOU