Dhanvantari GENOME TO HIT Genome Seq Protein 3





- Slides: 17

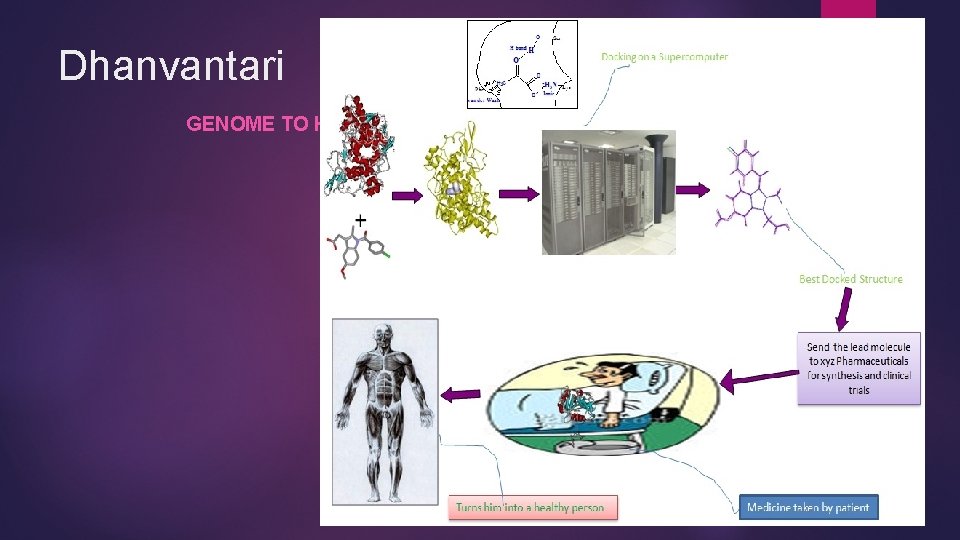
Dhanvantari GENOME TO HIT

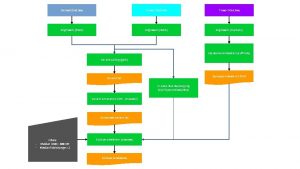
Genome Seq Protein 3 D Str Protein + Cavity Chemgenome A. . A Sequence Flowchart Bhageerath H / RM 2 TS+ Cavity known Prot. SAV/ Native RMSD/ Refine Str Yes ed iz m to ug us Dr AADS RASPD Method 1 C No Protein 3 D Str Database FDA, ZINC, NRDBSM, NCI or Customized Database RASPD Method 2 Par. DOCK Drug Structure ADMET Filters Final Leads

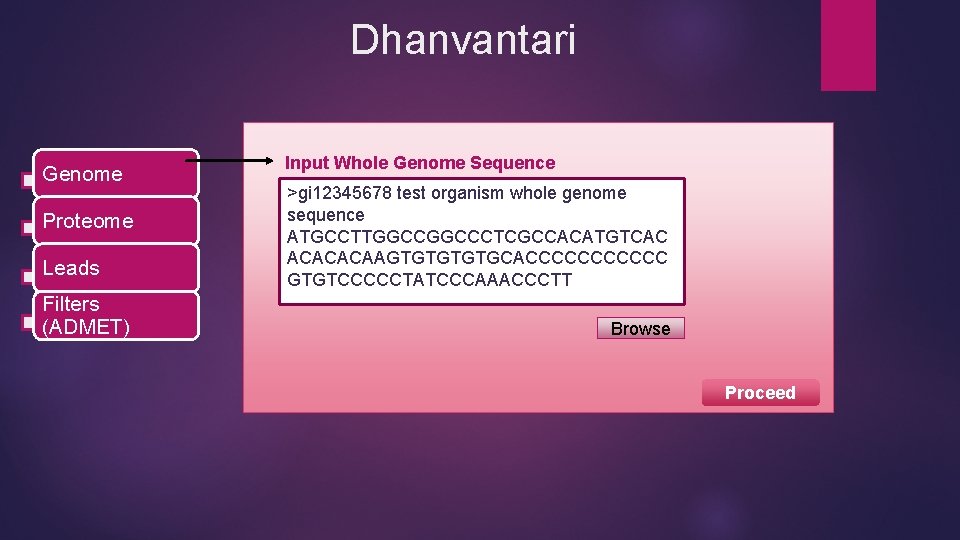
Dhanvantari Genome Proteome Leads Filters (ADMET) Input Whole Genome Sequence >gi 12345678 test organism whole genome sequence ATGCCTTGGCCCTCGCCACATGTCAC ACACACAAGTGTGCACCCCCC GTGTCCCCCTATCCCAAACCCTT Browse Proceed

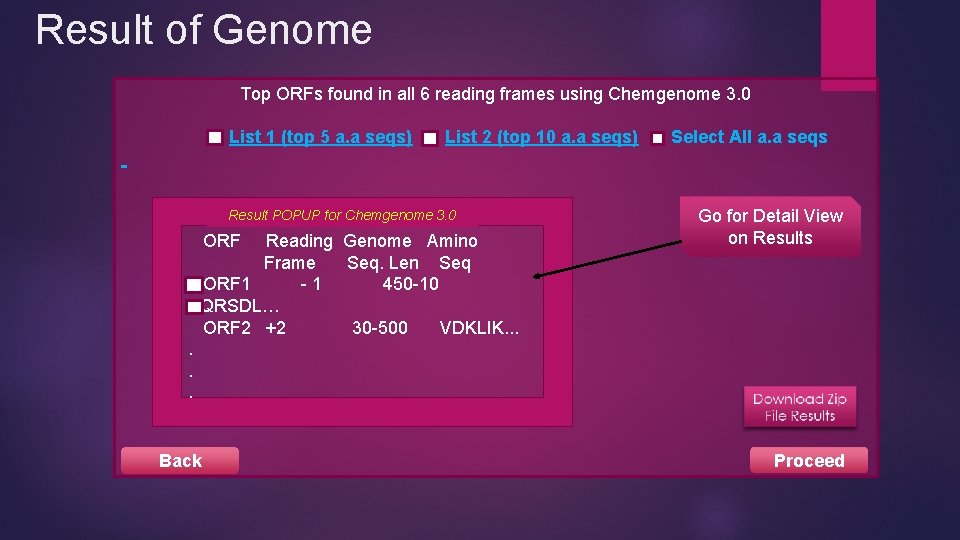
Result of Genome Top ORFs found in all 6 reading frames using Chemgenome 3. 0 List 1 (top 5 a. a seqs) List 2 (top 10 a. a seqs) Result POPUP for Chemgenome 3. 0 ORF Reading Genome Amino Frame Seq. Len Seq ORF 1 -1 450 -10 AQRSDL… ORF 2 +2 30 -500 VDKLIK. . . Back Select All a. a seqs Go for Detail View on Results Proceed

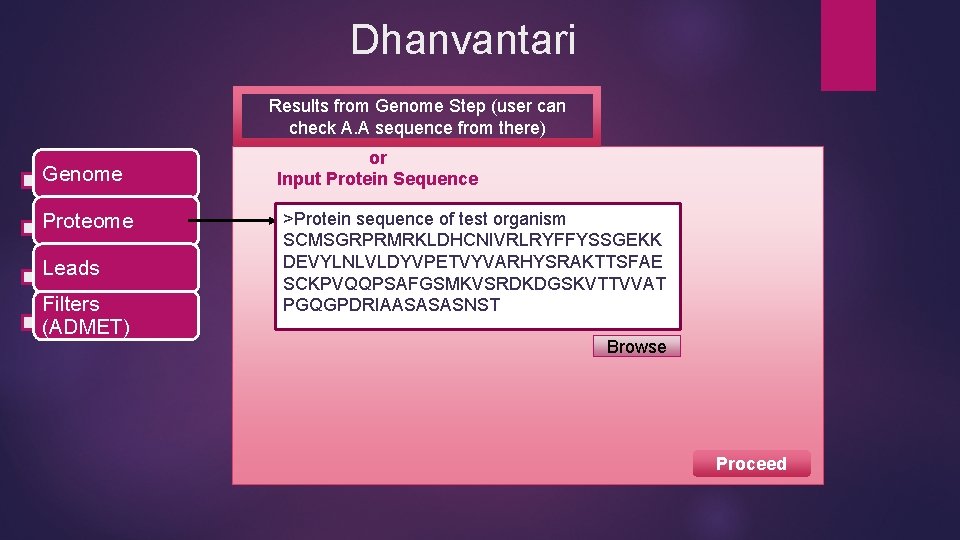
Dhanvantari Results from Genome Step (user can check A. A sequence from there) Genome Proteome Leads Filters (ADMET) or Input Protein Sequence >Protein sequence of test organism SCMSGRPRMRKLDHCNIVRLRYFFYSSGEKK DEVYLNLVLDYVPETVYVARHYSRAKTTSFAE SCKPVQQPSAFGSMKVSRDKDGSKVTTVVAT PGQGPDRIAASASASNST Browse Proceed

Result of Proteome Best Five Models built using BHAGEERATH H Model 1 Model 2 Model 3 Model 4 Result POPUP for Structure Evaluation Model Rank Model 1 1 Model 4 2. . . Protsav Score 3. 4 3. 9 Native RMSD 2. 9 A 3. 1 A Model 5 Select All Go for Structure Evaluation Refine Models Runs Refinery Program backend Download Zip File Results Back Proceed

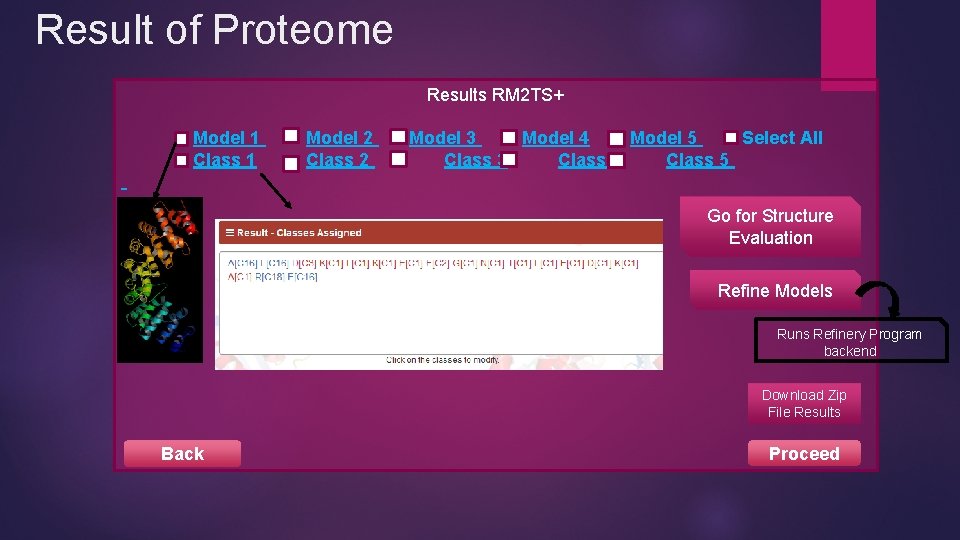
Result of Proteome Results RM 2 TS+ Model 1 Class 1 Model 2 Class 2 Model 3 Model 4 Model 5 Select All Class 3 Class 4 Class 5 Go for Structure Evaluation Refine Models Runs Refinery Program backend Download Zip File Results Back Proceed

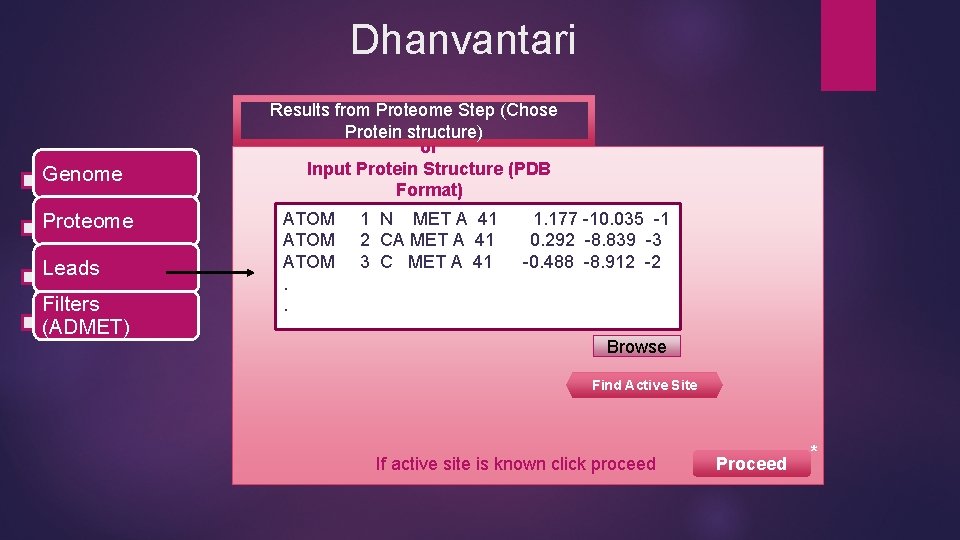
Dhanvantari Genome Proteome Leads Filters (ADMET) Results from Proteome Step (Chose Protein structure) or Input Protein Structure (PDB Format) ATOM. . 1 N MET A 41 2 CA MET A 41 3 C MET A 41 1. 177 -10. 035 -1 0. 292 -8. 839 -3 -0. 488 -8. 912 -2 Browse Find Active Site If active site is known click proceed Proceed *

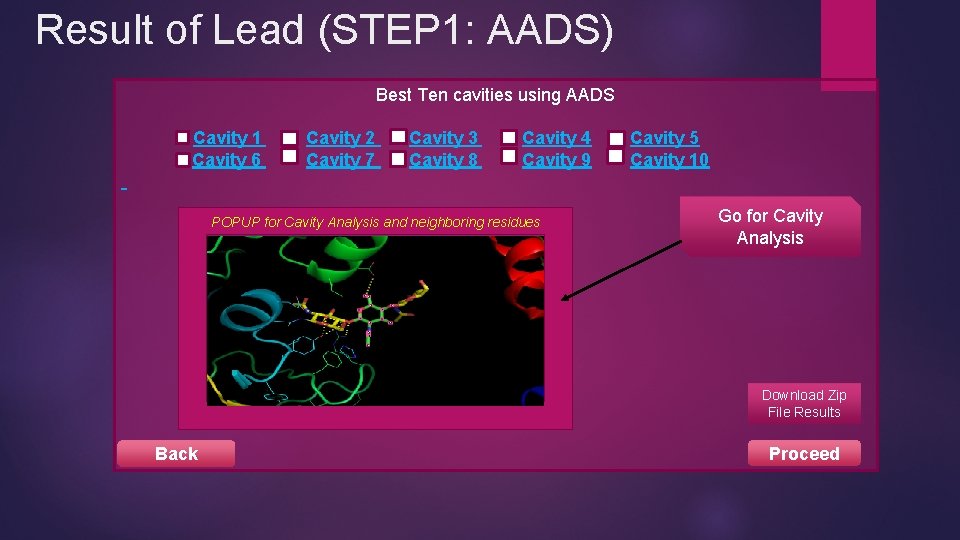
Result of Lead (STEP 1: AADS) Best Ten cavities using AADS Cavity 1 Cavity 6 Cavity 2 Cavity 7 Cavity 3 Cavity 8 Cavity 4 Cavity 9 POPUP for Cavity Analysis and neighboring residues Cavity 5 Cavity 10 Go for Cavity Analysis Download Zip File Results Back Proceed

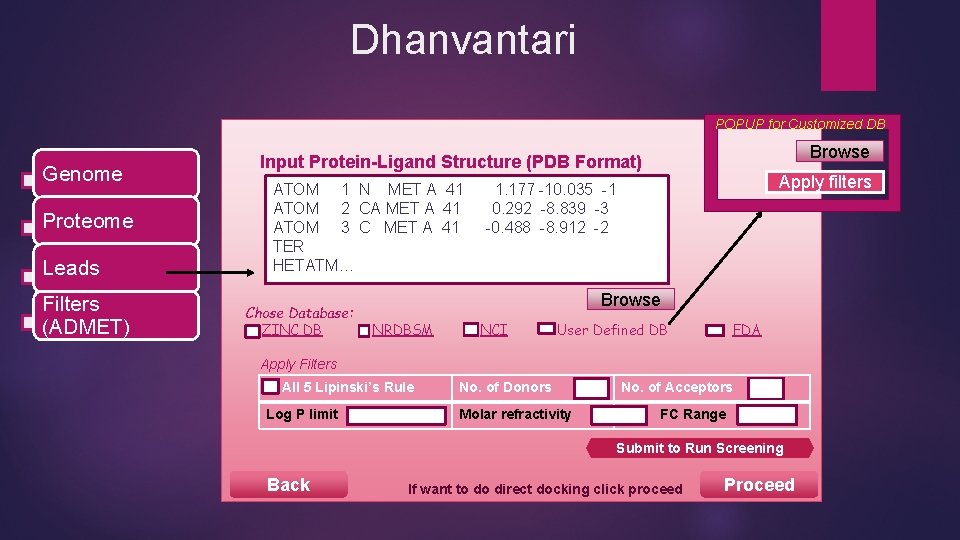
Dhanvantari POPUP for Customized DB Genome Proteome Leads Filters (ADMET) Browse Input Protein-Ligand Structure (PDB Format) ATOM 1 N MET A 41 ATOM 2 CA MET A 41 ATOM 3 C MET A 41 TER HETATM… Chose Database: ZINC DB Apply filters 1. 177 -10. 035 -1 0. 292 -8. 839 -3 -0. 488 -8. 912 -2 Browse NRDBSM NCI User Defined DB FDA Apply Filters All 5 Lipinski’s Rule Log P limit No. of Donors Molar refractivity No. of Acceptors FC Range Submit to Run Screening Back If want to do direct docking click proceed Proceed

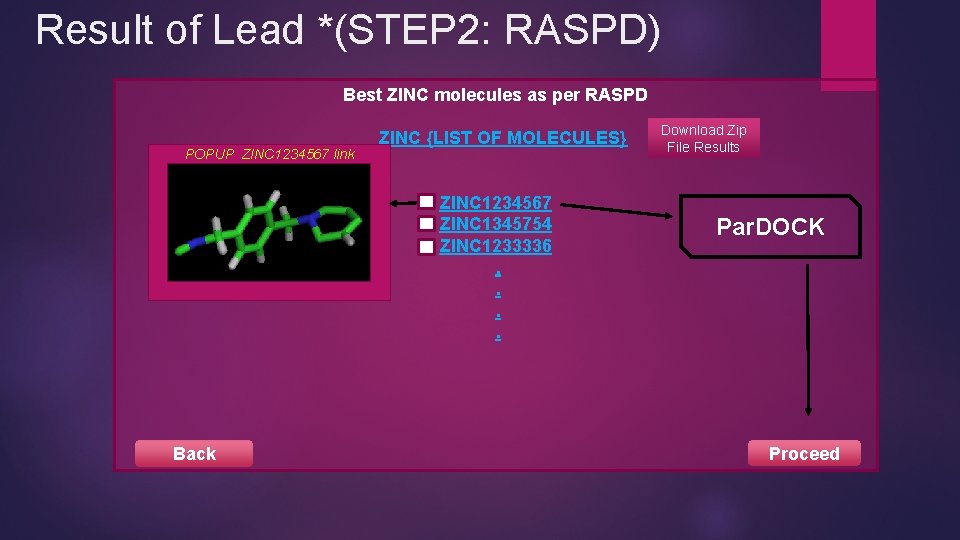
Result of Lead *(STEP 2: RASPD) Best ZINC molecules as per RASPD POPUP ZINC 1234567 link ZINC {LIST OF MOLECULES} ZINC 1234567 ZINC 1345754 ZINC 1233336. . Back Download Zip File Results Par. DOCK Proceed

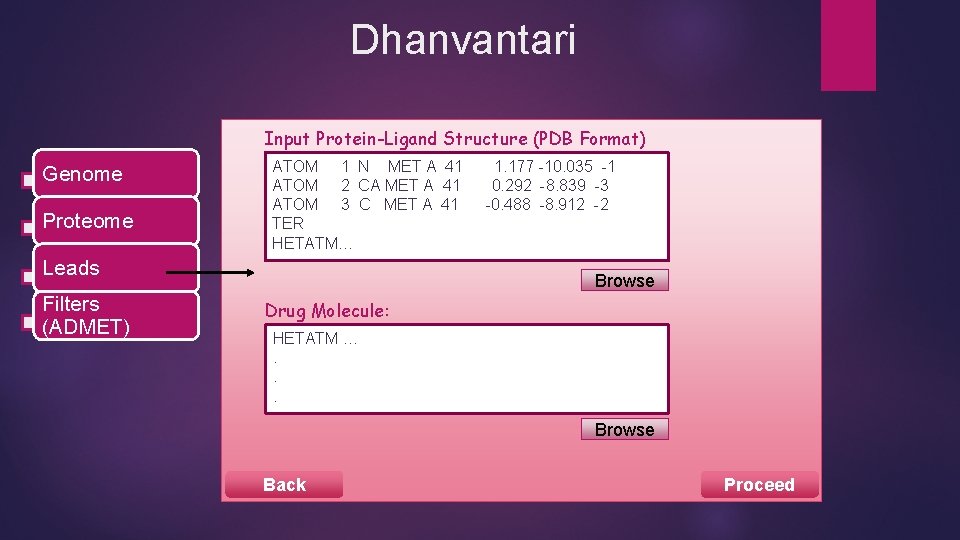
Dhanvantari Input Protein-Ligand Structure (PDB Format) Genome Proteome ATOM 1 N MET A 41 ATOM 2 CA MET A 41 ATOM 3 C MET A 41 TER HETATM… Leads Filters (ADMET) 1. 177 -10. 035 -1 0. 292 -8. 839 -3 -0. 488 -8. 912 -2 Browse Drug Molecule: HETATM …. . . Browse Back Proceed

Result of Lead Step 3 (Par. DOCK) Docking Score based on Binding Energy calculated by Par. DOCK Proposed Lead molecules with Binding Poses POPUP ZINC 1234567 link ZINC 1234567 Energy 1 ZINC 1345754 Energy 2 ZINC 1233336 Energy 3. . . Back Pose 1 Pose 2 Pose 3 Pose 4 POPUP Pose 1 View Proceed

An additional ADMET calculator could be added

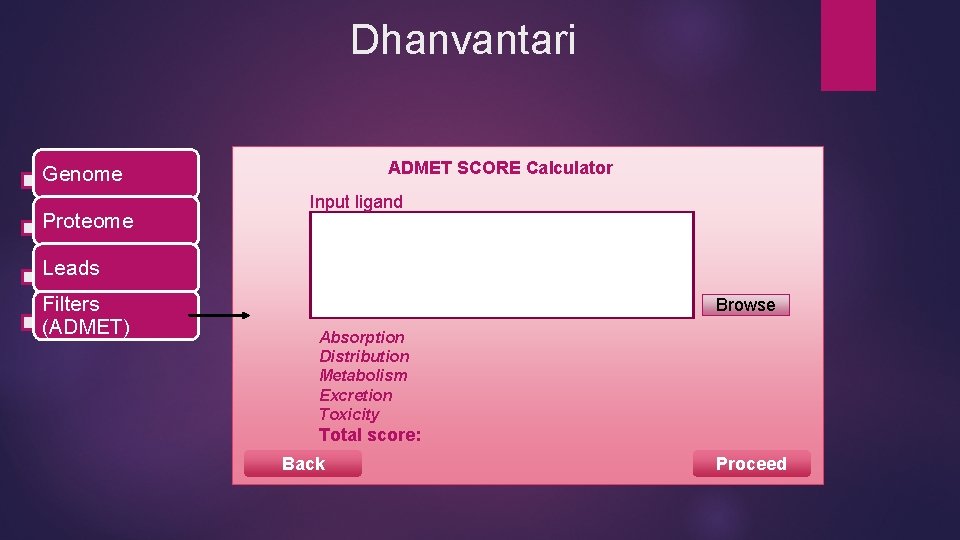
Dhanvantari ADMET SCORE Calculator Genome Proteome Input ligand Leads Filters (ADMET) Browse Absorption Distribution Metabolism Excretion Toxicity Total score: Back Proceed

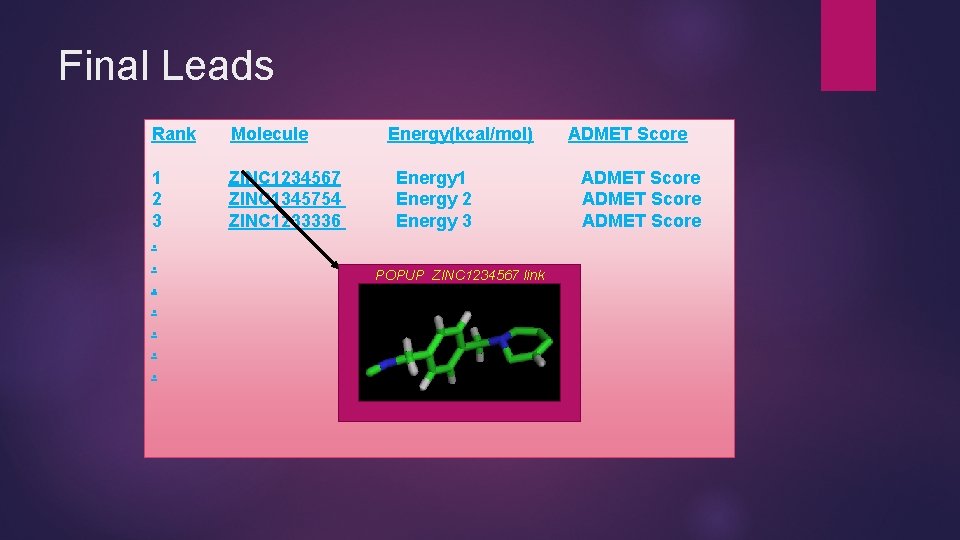
Final Leads Rank Molecule 1 2 3. . . . ZINC 1234567 ZINC 1345754 ZINC 1233336 Energy(kcal/mol) Energy 1 Energy 2 Energy 3 POPUP ZINC 1234567 link ADMET Score

Thank you
 Semi-global alignment
Semi-global alignment Slide-seq
Slide-seq Per base sequence quality
Per base sequence quality Reversible terminator
Reversible terminator Seq monk
Seq monk Tcp sequence diagram
Tcp sequence diagram Struktur sequence
Struktur sequence Seq artinya
Seq artinya Ocaml seq
Ocaml seq Tuxedo suite rna seq
Tuxedo suite rna seq Composite structure diagram
Composite structure diagram Gan chatbot
Gan chatbot Channel vs carrier proteins
Channel vs carrier proteins Protein-protein docking
Protein-protein docking What happened to light when it hits an object
What happened to light when it hits an object Hit the button
Hit the button Multi hit model
Multi hit model They were ___ talented singers that the concert was a hit
They were ___ talented singers that the concert was a hit