Developmental Integration of Bioinformatics Activities at Different Levels
Developmental Integration of Bioinformatics Activities at Different Levels of the Biology Curriculum Jeff Newman Lycoming College, Williamsport PA August 5, 2016
Genomics & Bioinformatics Throughout the Curriculum Jeffrey D. Newman, Lycoming College November 10, 2006

Outline • The starting line: Where we were. • Philosophy: Use of bioinformatics & genome data is as important to a 21 st century biologist as using a microscope!!! • 3 phases – Incorporate Molecular Biology, Incorporate Genomics and Bioinformatics, Add New Upper-Level Courses. – Introductory Biology – Genetics – Microbiology – Upper-level courses – Biochemistry, Molecular Biology, Genome Analysis, Cell & Molecular Research Methods. • Assessment Surveys – Knowledge, Skills, Attitudes • Where to go from here?
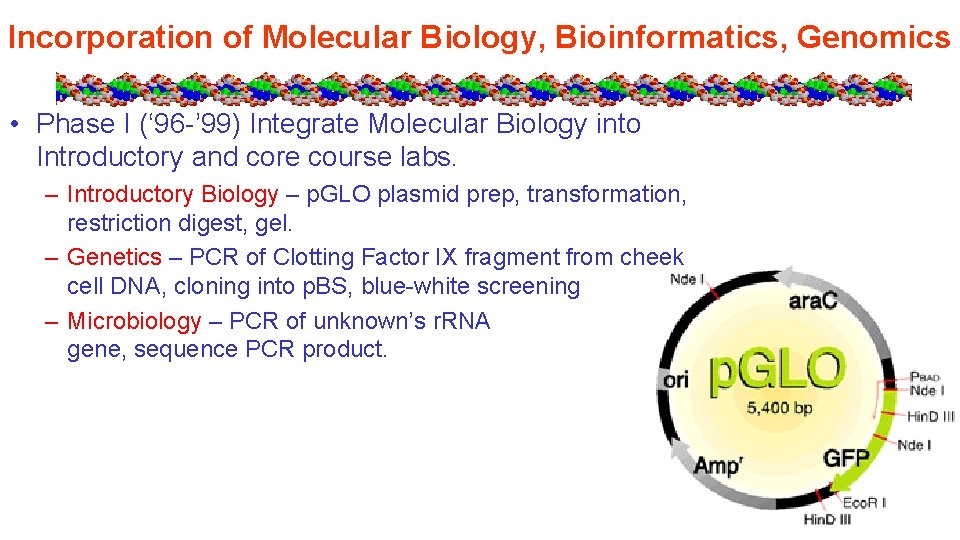
Incorporation of Molecular Biology, Bioinformatics, Genomics • Phase I (‘ 96 -’ 99) Integrate Molecular Biology into Introductory and core course labs. – Introductory Biology – p. GLO plasmid prep, transformation, restriction digest, gel. – Genetics – PCR of Clotting Factor IX fragment from cheek cell DNA, cloning into p. BS, blue-white screening – Microbiology – PCR of unknown’s r. RNA gene, sequence PCR product.
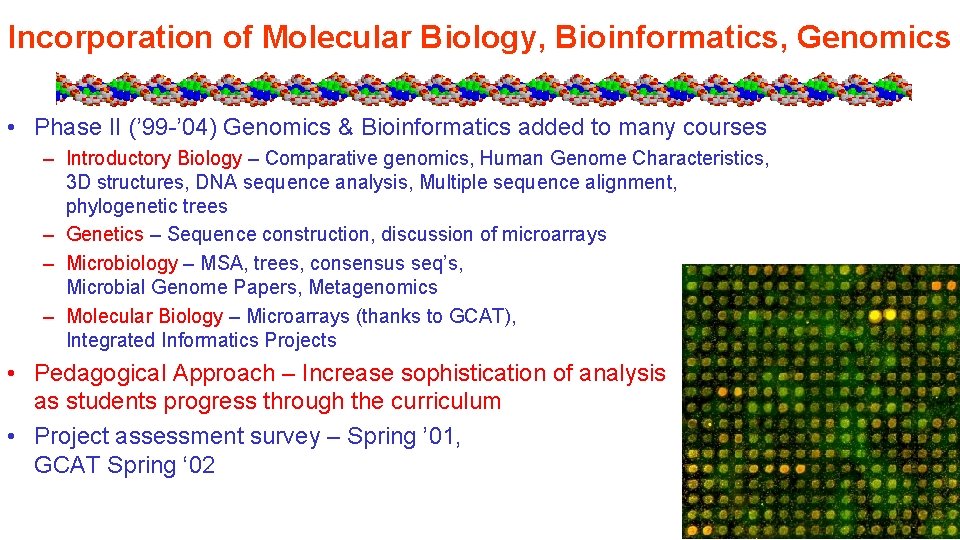
Incorporation of Molecular Biology, Bioinformatics, Genomics • Phase II (’ 99 -’ 04) Genomics & Bioinformatics added to many courses – Introductory Biology – Comparative genomics, Human Genome Characteristics, 3 D structures, DNA sequence analysis, Multiple sequence alignment, phylogenetic trees – Genetics – Sequence construction, discussion of microarrays – Microbiology – MSA, trees, consensus seq’s, Microbial Genome Papers, Metagenomics – Molecular Biology – Microarrays (thanks to GCAT), Integrated Informatics Projects • Pedagogical Approach – Increase sophistication of analysis as students progress through the curriculum • Project assessment survey – Spring ’ 01, GCAT Spring ‘ 02
Incorporation of Molecular Biology, Bioinformatics, Genomics • Phase III (’ 04 - ? ) – New course development – Genome Analysis – Fall ’ 04, ‘ 06 – Cell and Molecular Research Methods, Fall ‘ 06

Courses Taught (all have labs except Public Health) • Fall – Bio 110–Introduction to Biology I (with 2 -3 lab sections) – Bio 150–Public Health or Bio 432–Molecular Biology or Bio 437 – Genome Analysis or Bio 447 – Research Methods • Spring – Bio 321 – Microbiology (with 2 lab sections) – Bio/Chem 444 – Biochemistry • Research lab with 5 -15 students – – Research Methods, Independent Study & Honors students, Paid lab assistants High school student volunteers
Bio 110 – Introduction to Biology I (majors) Lab activities designed to support course topics. • • Biomolecules – Lab #2 = 3 D structures of molecules Cell Biology Enzymes & Metabolism – Lab #4 c = Kinetic Analysis with Excel Information Flow – Lab #5 a = Gene ID in a sequence, predicting traits from plasmid and genome, restriction mapping of plasmid. Cell signaling Cell Cycle Mutations Cancer Meiosis Mendelian Genetics – Lab #7 = OMIM for basis of traits Biotech, Genomics, Developmental Biology Evolution, Population Genetics – Lab #10 = Retrieve myoglobin protein sequences from different animals, align, create tree, ID lineages where mutations occurred.

Intro Bio Lab #2 = 3 D structures of molecules • Small Molecules using Biomodel-3, developed by Angel Herráez (angel. herraez@uah. es), lecturer in Biochemistry and Molecular Biology at the University of Alcalá de Henares (Spain). http: //biomodel. uah. es/en/model 3/inicio. htm • Concepts – pdb files, rendering structures in different ways, manipulating structures, standard color schemes for elements. – # of bonds on atoms, chemical formula, atomic/molecular mass, functional groups – Saturated vs unsaturated fatty acids, components of phospholipids, arrangement of phospholipids into a bilayer
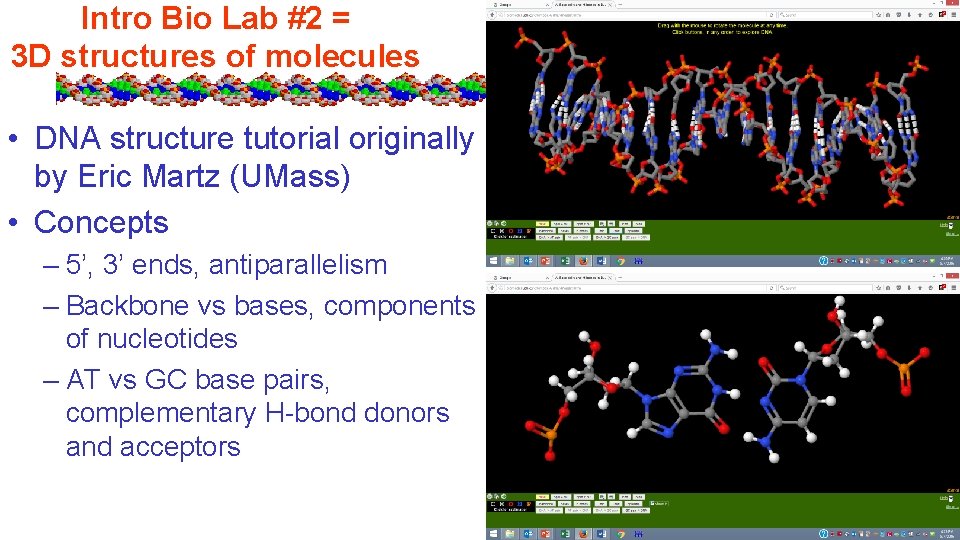
Intro Bio Lab #2 = 3 D structures of molecules • DNA structure tutorial originally by Eric Martz (UMass) • Concepts – 5’, 3’ ends, antiparallelism – Backbone vs bases, components of nucleotides – AT vs GC base pairs, complementary H-bond donors and acceptors
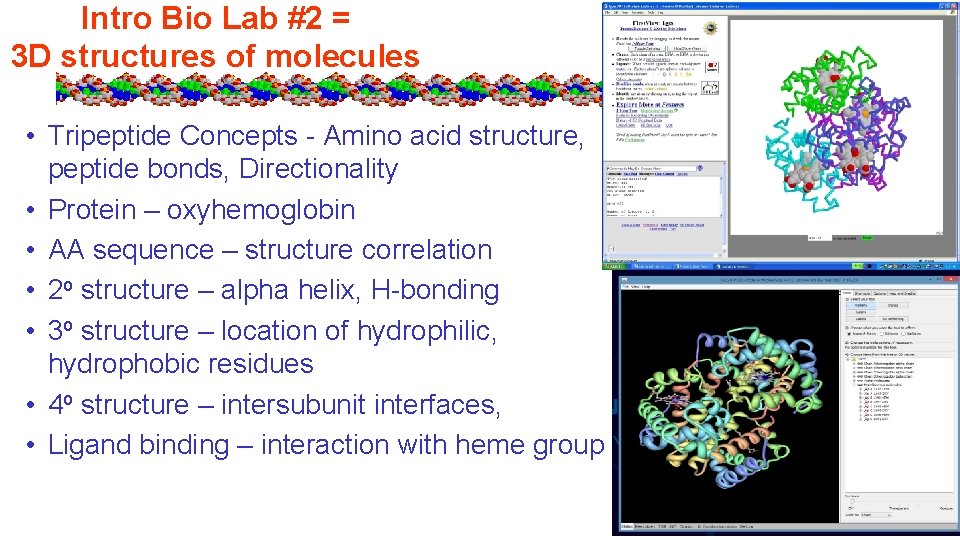
Intro Bio Lab #2 = 3 D structures of molecules • Tripeptide Concepts - Amino acid structure, peptide bonds, Directionality • Protein – oxyhemoglobin • AA sequence – structure correlation • 2 o structure – alpha helix, H-bonding • 3 o structure – location of hydrophilic, hydrophobic residues • 4 o structure – intersubunit interfaces, • Ligand binding – interaction with heme group
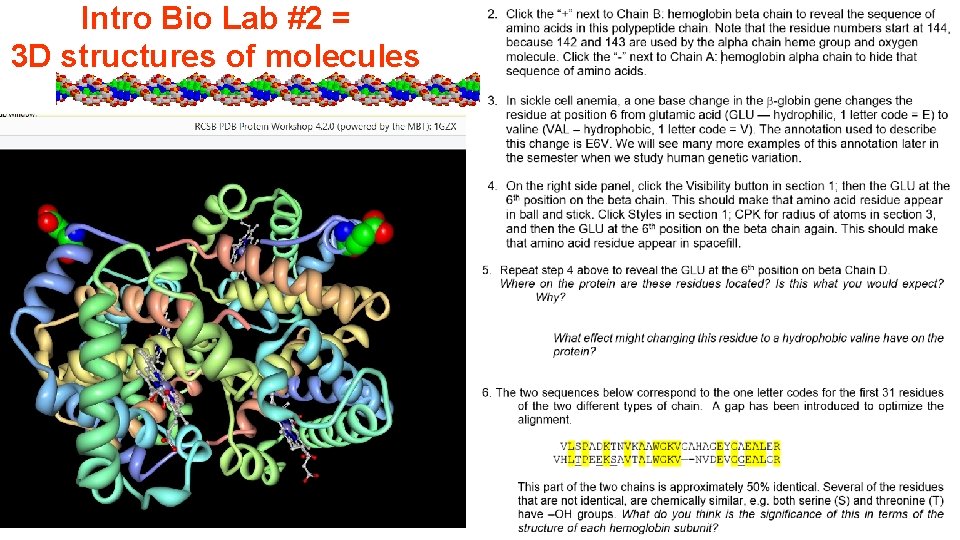
Intro Bio Lab #2 = 3 D structures of molecules
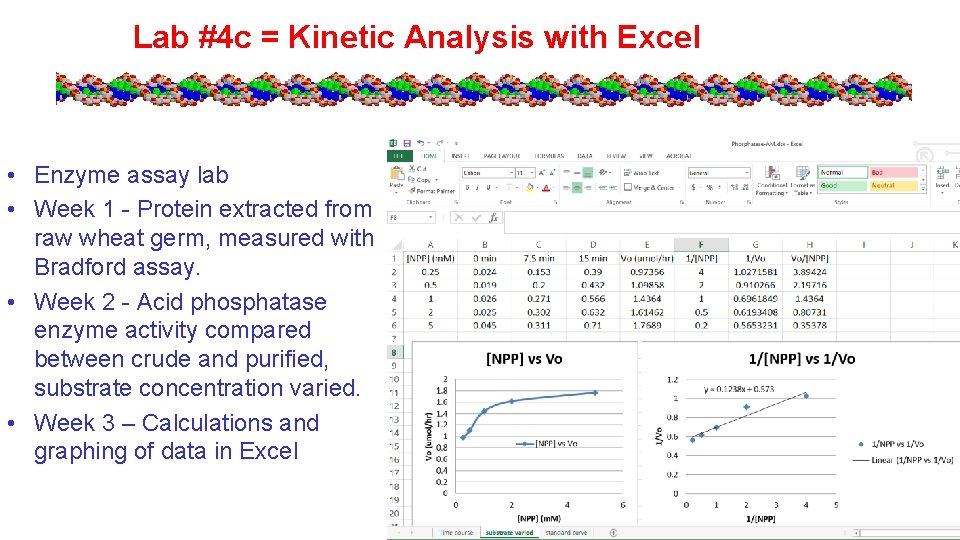
Lab #4 c = Kinetic Analysis with Excel • Enzyme assay lab • Week 1 - Protein extracted from raw wheat germ, measured with Bradford assay. • Week 2 - Acid phosphatase enzyme activity compared between crude and purified, substrate concentration varied. • Week 3 – Calculations and graphing of data in Excel
Lab #5 a = Mr. Green Genes - Gene ID in a sequence, predicting traits from plasmid and genome, restriction mapping of plasmid. • Lab developed using Bio. Rad’s p. GLO plasmid. • Students provided with p. GLO DNA sequence find genes develop hypotheses about traits of bacteria with plasmid design experiments to test hypothesis about function of the DNA find restriction sites develop hypotheses/predictions about fragment sizes after cutting with restriction enzyme tests physical properties of the DNA
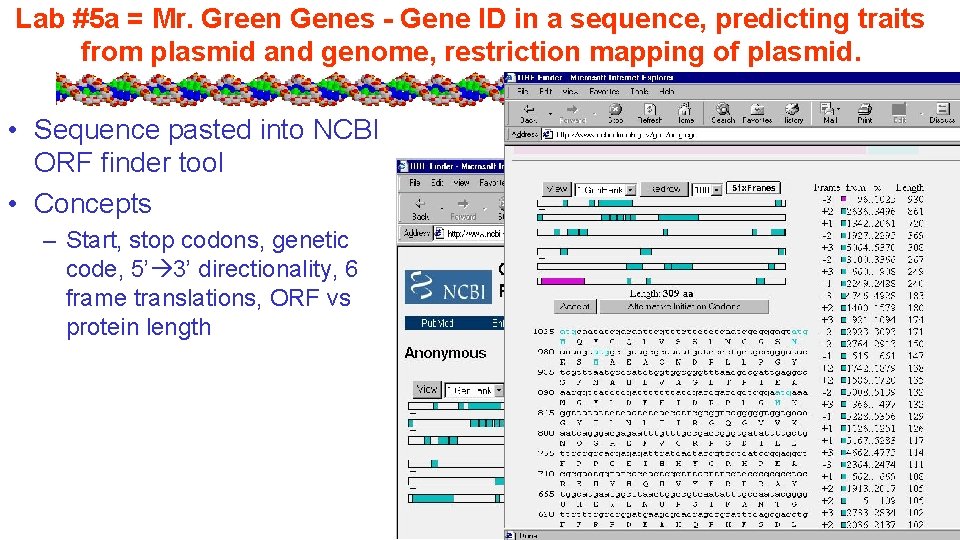
Lab #5 a = Mr. Green Genes - Gene ID in a sequence, predicting traits from plasmid and genome, restriction mapping of plasmid. • Sequence pasted into NCBI ORF finder tool • Concepts – Start, stop codons, genetic code, 5’ 3’ directionality, 6 frame translations, ORF vs protein length
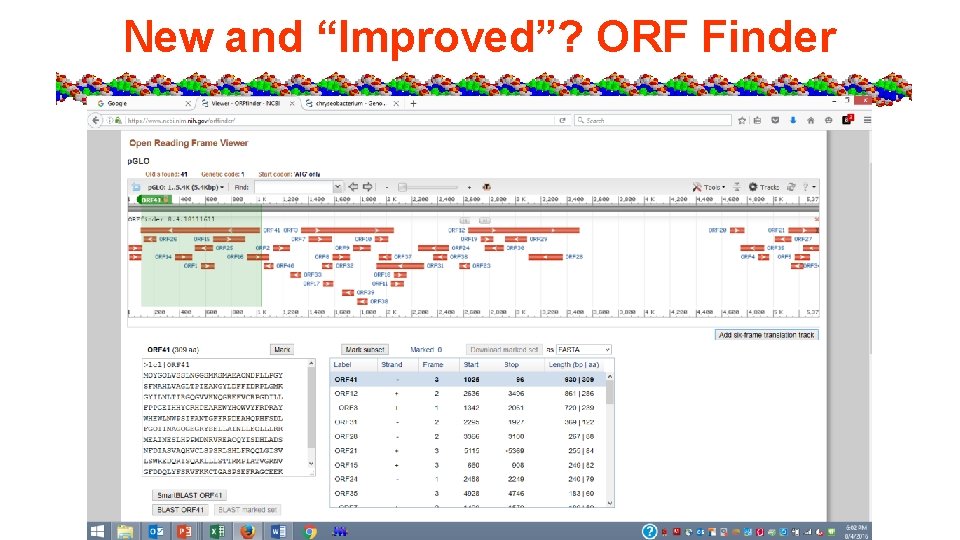
New and “Improved”? ORF Finder
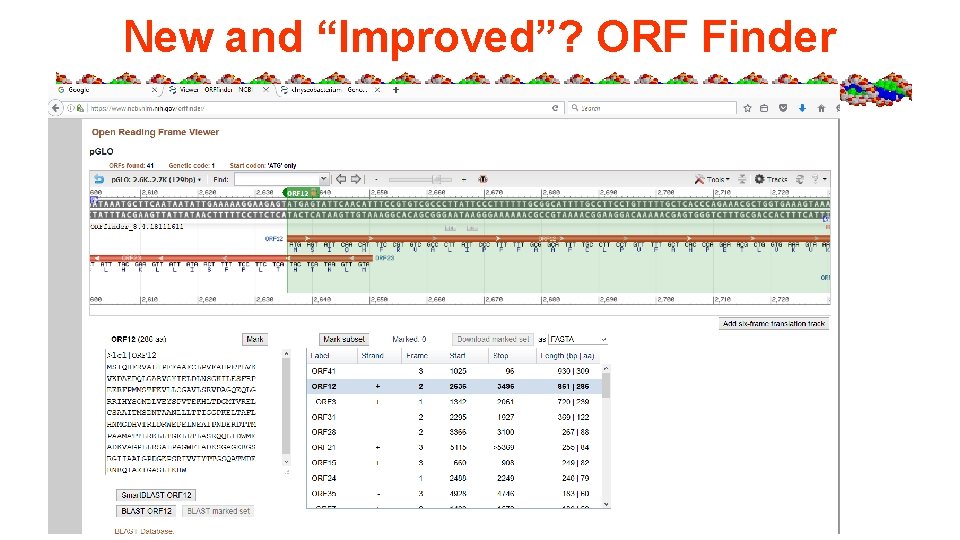
New and “Improved”? ORF Finder
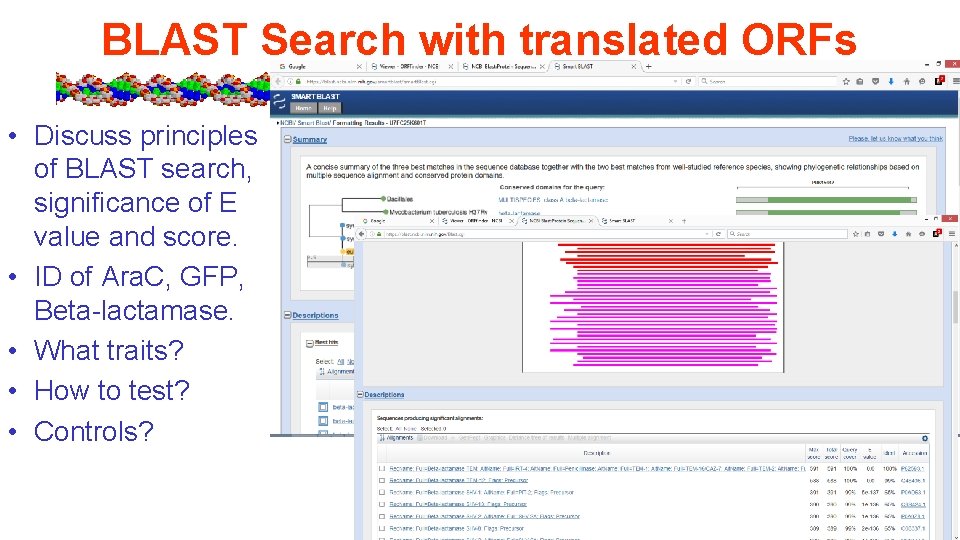
BLAST Search with translated ORFs • Discuss principles of BLAST search, significance of E value and score. • ID of Ara. C, GFP, Beta-lactamase. • What traits? • How to test? • Controls?
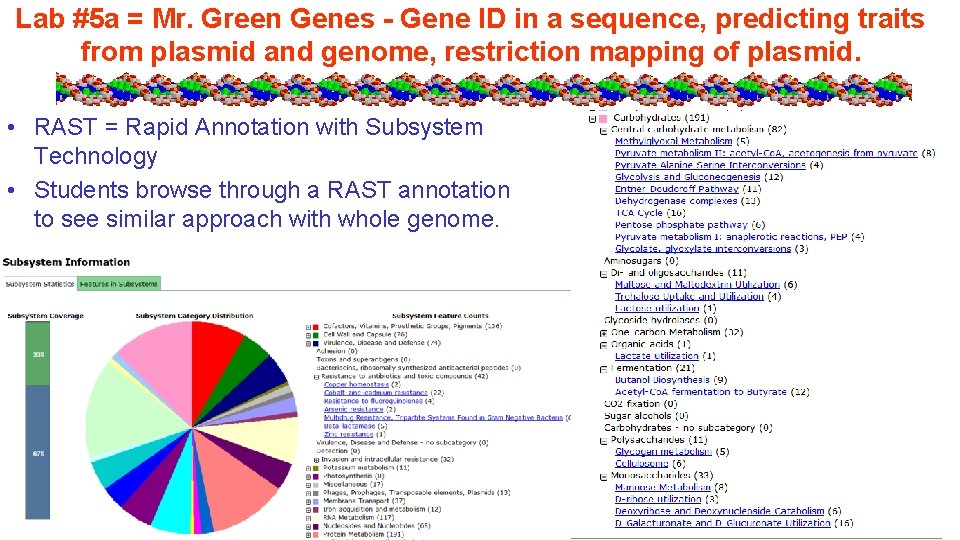
Lab #5 a = Mr. Green Genes - Gene ID in a sequence, predicting traits from plasmid and genome, restriction mapping of plasmid. • RAST = Rapid Annotation with Subsystem Technology • Students browse through a RAST annotation to see similar approach with whole genome.
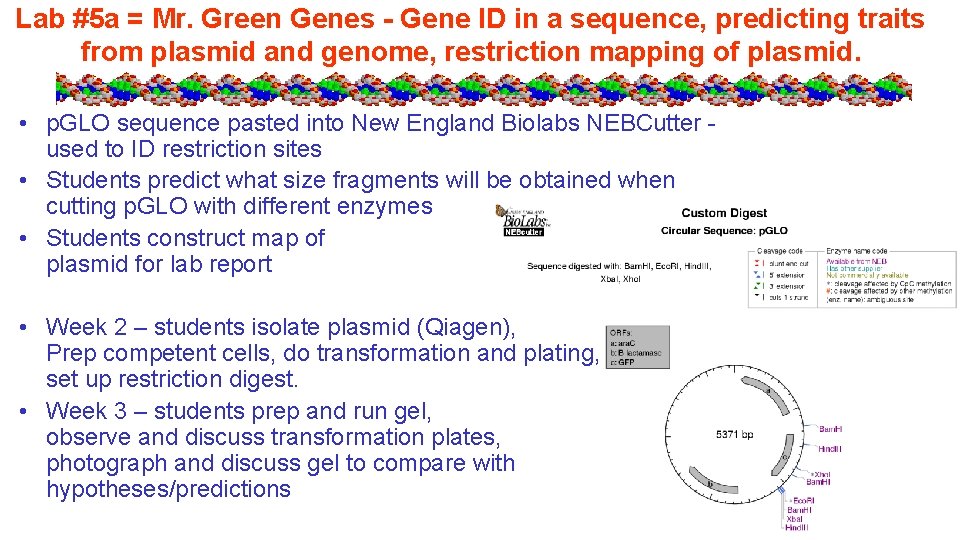
Lab #5 a = Mr. Green Genes - Gene ID in a sequence, predicting traits from plasmid and genome, restriction mapping of plasmid. • p. GLO sequence pasted into New England Biolabs NEBCutter used to ID restriction sites • Students predict what size fragments will be obtained when cutting p. GLO with different enzymes • Students construct map of plasmid for lab report • Week 2 – students isolate plasmid (Qiagen), Prep competent cells, do transformation and plating, set up restriction digest. • Week 3 – students prep and run gel, observe and discuss transformation plates, photograph and discuss gel to compare with hypotheses/predictions
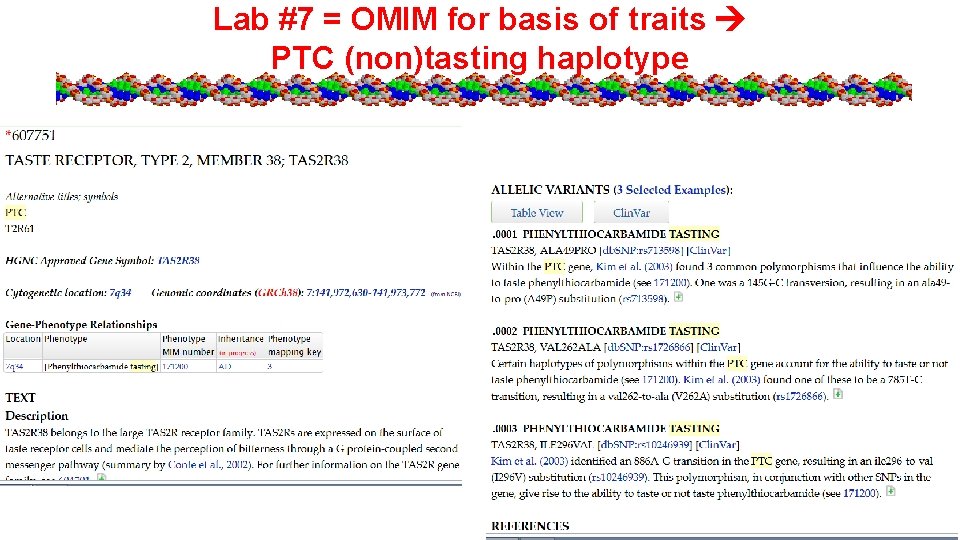
Lab #7 = OMIM for basis of traits PTC (non)tasting haplotype
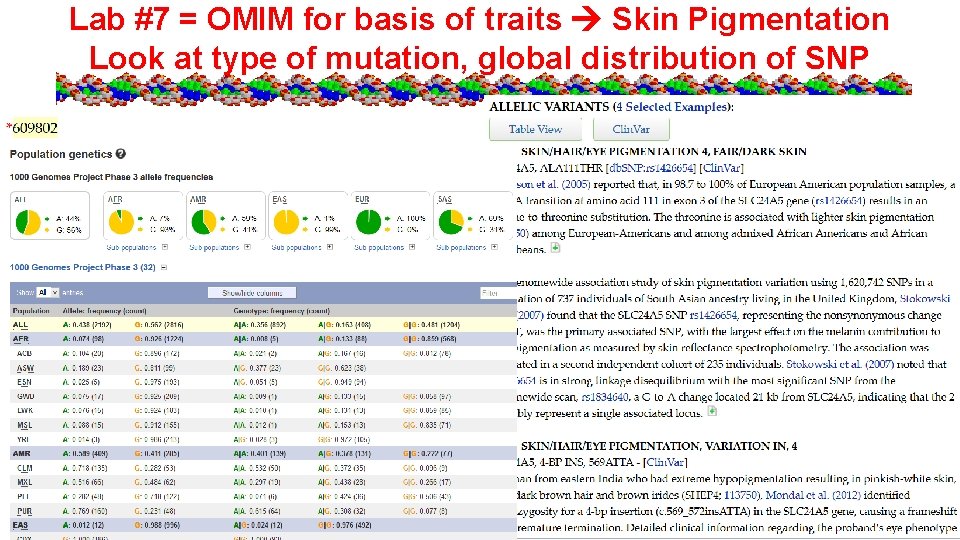
Lab #7 = OMIM for basis of traits Skin Pigmentation Look at type of mutation, global distribution of SNP

Lab #7 = OMIM for basis of traits Red Hair/Fair Skin Multiple genes, phenotypes, signals, tanning response
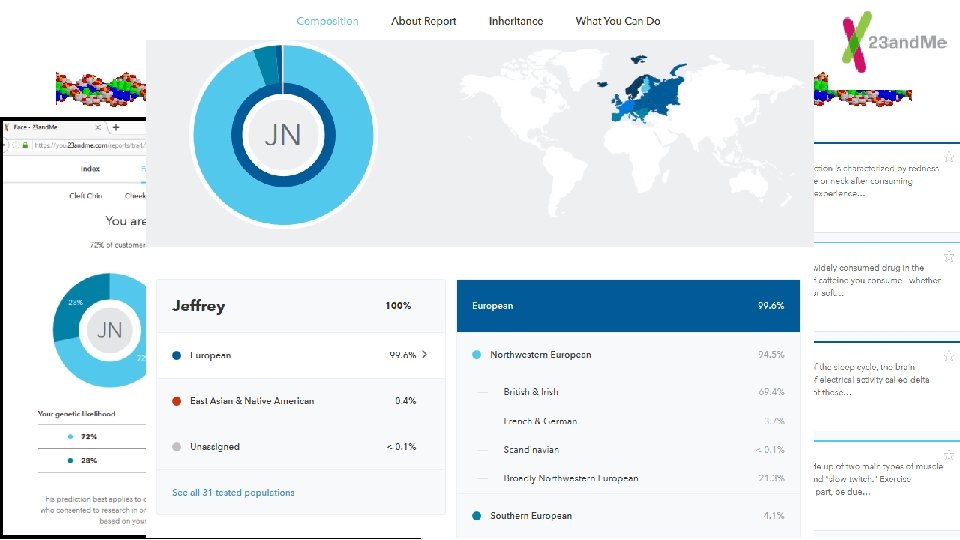
Lab #7 = OMIM for basis of traits Colorblindness, Blood type…. 23 and Me
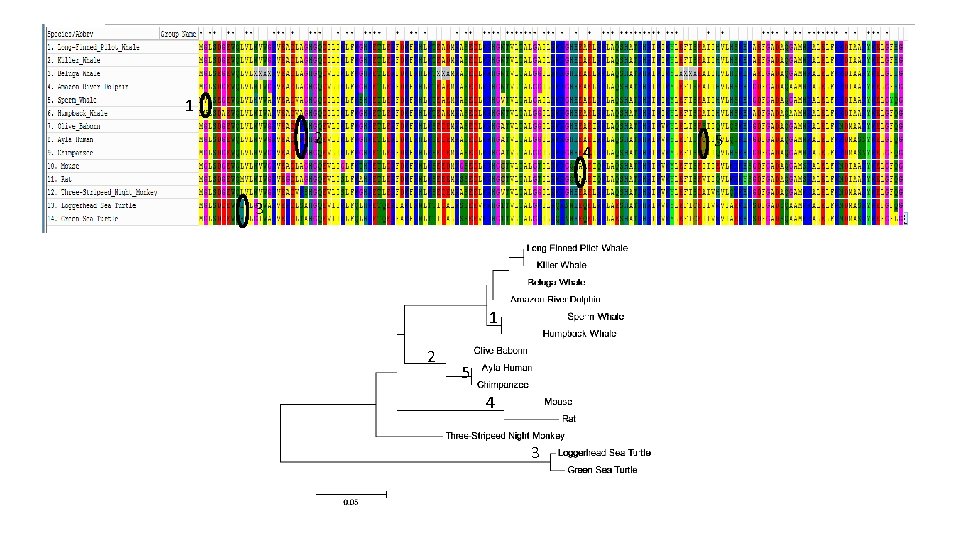
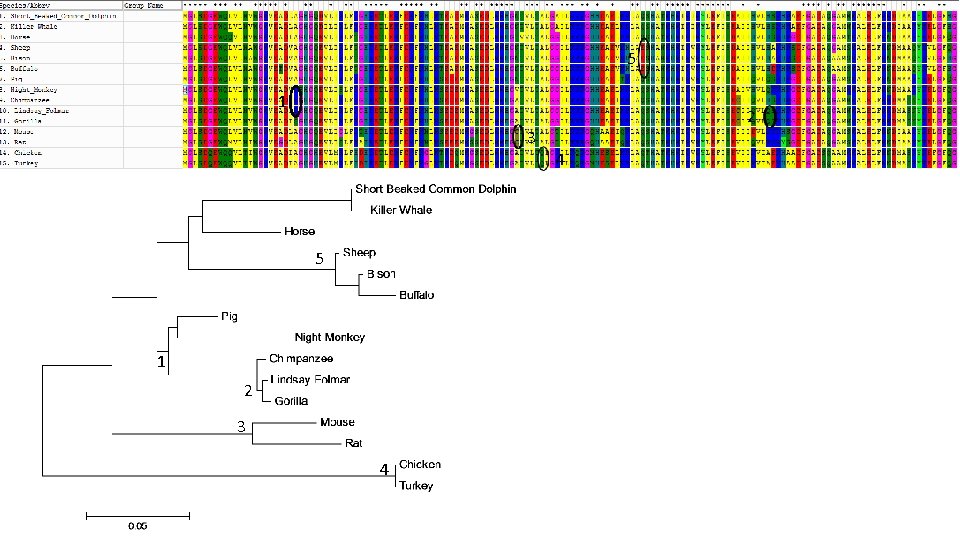
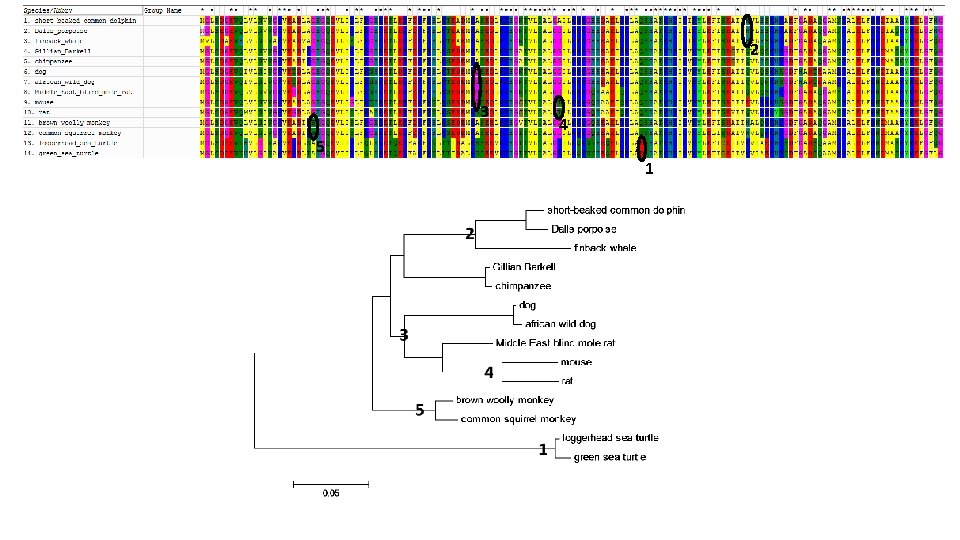
Lab #10 = Retrieve myoglobin protein sequences from different animals, align, create tree, ID mutations. • Week 14 lab – Hybrid lab on Evolution – Watch video, compare hominid skulls, ape chromosome banding patterns, myoglobin sequences. • Uni. Prot used to retrieve myoglobin protein sequences from a diverse set of vertebrates, including Human • After editing species names, MEGA used to create multiple sequence alignment & construct phylogenetic tree • Students compare tree and alignment to ID ancestor where mutations occurred. • Best evidence for evolution
1 2 4 3 1 2 5 4 3 5
5 1 3 5 1 2 3 4 2 4
2 3 5 4 1
Bioinformatics in Intro Biology - Summary • Students spend 4. 5 lab periods in the computer lab • Advantages – Students develop key skills, become experienced with basic bioinformatics tools and databases – Abstract concepts become more concrete through hands-on analysis and visualization – It’s free!!!! • Disadvantages – Fewer wet labs, frequent software and web site changes require regular revision of instructions – Some students find computer work boring
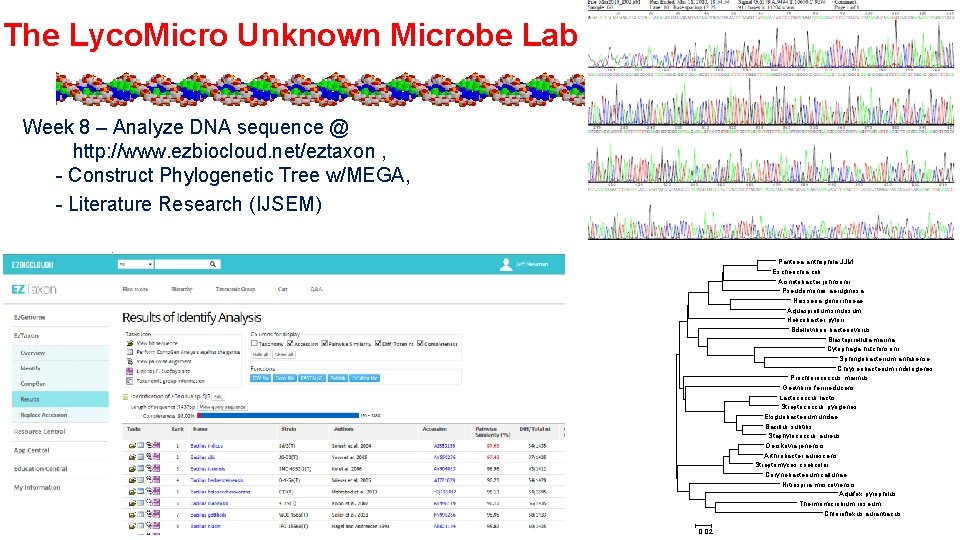
The Lyco. Micro Unknown Microbe Lab Week 8 – Analyze DNA sequence @ http: //www. ezbiocloud. net/eztaxon , - Construct Phylogenetic Tree w/MEGA, - Literature Research (IJSEM) Pantoea anthophila JJM Escherichia coli Acinetobacter johnsonii Pseudomonas aeruginosa Neisseria gonorrhoeae Aquaspirillum sinuosum Helicobacter pylori Bdellovibrio bacteriovorus Blastopirellula marina Cytophaga hutchinsonii Sphingobacterium anhuiense Chryseobacterium indologenes Prochlorococcus marinus Geovibrio ferrireducens Lactococcus lactis Streptococcus pyogenes Exiguobacterium undae Bacillus subtilis Staphylococcus aureus Oerskovia jenensis Arthrobacter aurescens Streptomyces coelicolor Corynebacterium callunae Nitrospira moscoviensis Aquifex pyrophilus Thermomicrobium roseum Chloroflexus aurantiacus 0. 02
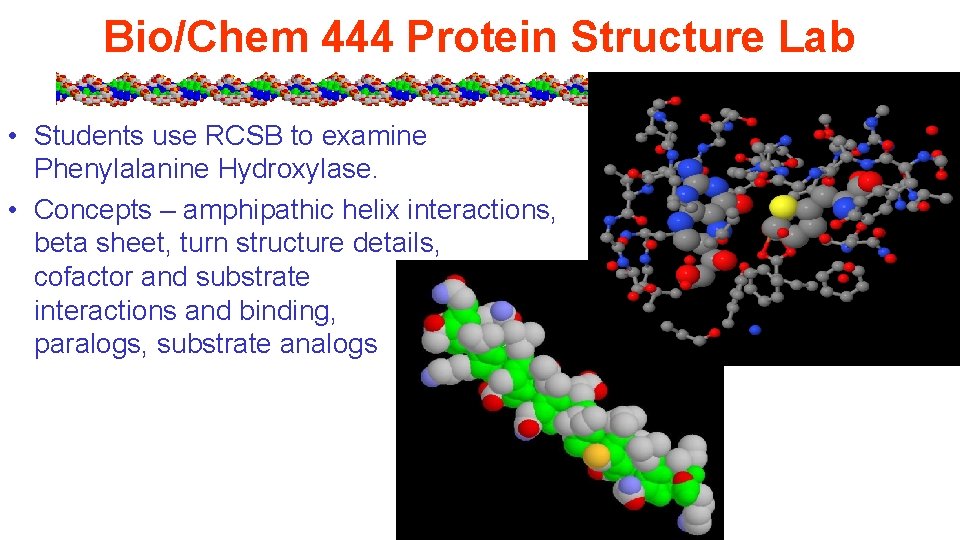
Bio/Chem 444 Protein Structure Lab • Students use RCSB to examine Phenylalanine Hydroxylase. • Concepts – amphipathic helix interactions, beta sheet, turn structure details, cofactor and substrate interactions and binding, paralogs, substrate analogs
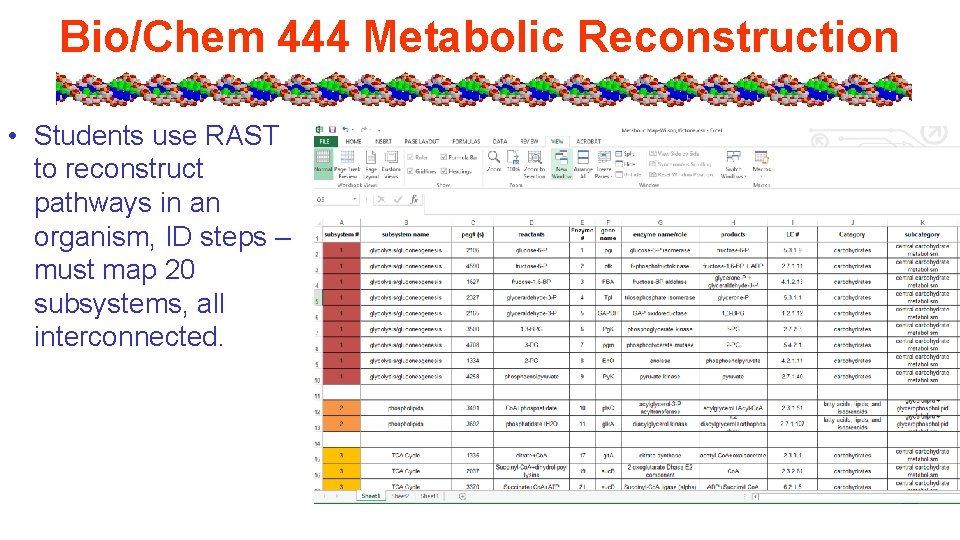
Bio/Chem 444 Metabolic Reconstruction • Students use RAST to reconstruct pathways in an organism, ID steps – must map 20 subsystems, all interconnected.
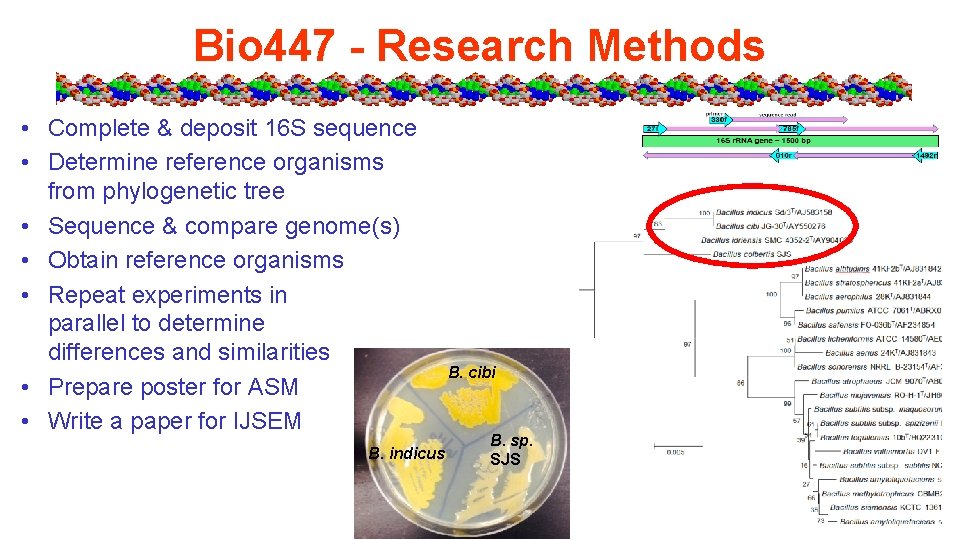
Bio 447 - Research Methods • Complete & deposit 16 S sequence • Determine reference organisms from phylogenetic tree • Sequence & compare genome(s) • Obtain reference organisms • Repeat experiments in parallel to determine differences and similarities • Prepare poster for ASM • Write a paper for IJSEM B. indicus B. cibi B. sp. SJS
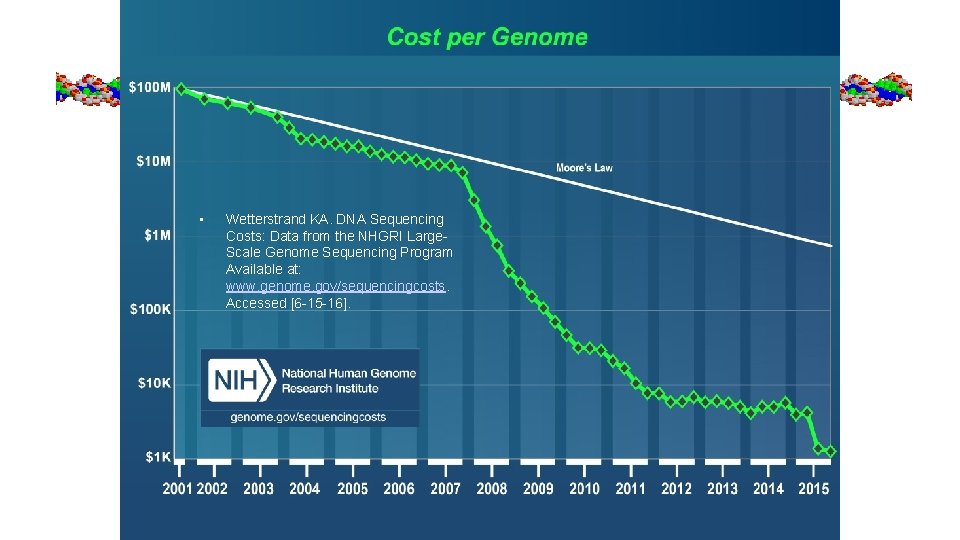
• Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Large. Scale Genome Sequencing Program Available at: www. genome. gov/sequencingcosts. Accessed [6 -15 -16].

GCAT-SEEK • Genome Consortium for Active Teaching (GCAT) founded in 2000 to bring Genomics (Microarrays) to the undergraduate curriculum. • Multiple HHMI & NSF funded workshops • GCAT-SEEKquence “spin-off” to bring Next. Gen sequencing to the undergraduate curriculum. • 3 genomes (Ion Torrent & 454 as part of pilot) • NSF Research Collaboration Network, Juniata’s HHMI Genomics Leadership Initiative

Shared Mi. Seq (2 x 300) Runs • Next. Gen Instruments generate more data than most UG faculty can use or afford. • November 2013 – 27 bacteria @$200 each (including Flavobacterium aquatile) • April, 2014 – Opened to Microedu Listserv 35 Bacteria and Phage from 16 institutions @$190/sample • October 2014 – 30 phage, viruses and bacteria @$175/sample. Sample Reads est. Bases est. GSF 665 -1 -E_coli-C 06 b GSF 665 -2 -Chryseobacterium-LO GSF 665 -3 -Linfield-KH GSF 665 -4 -Linfield-NH GSF 665 -5 -Exiguobacterium GSF 665 -6 -Plesiomonas_shigelloides GSF 665 -7 -Halosimplex_carlsbadense GSF 665 -8 -Phage_Eapen GSF 665 -9 -Phage_Aspire GSF 665 -10 -strain_3572 GSF 665 -11 -Gracilibacillus_dipsosauri GSF 665 -12 -Serratia_S 12 GSF 665 -13 -Rhodococcus_T 1 Sofl-14 GSF 665 -14 -Janthinobacterium-BJB 1 GSF 665 -15 -Janthinobacterium-BJB 349 GSF 665 -16 -Janthinobacterium-BJB 304 GSF 665 -17 -Janthinobacterium-BJB 317 GSF 665 -18 -Iodobacter-BJB 302 GSF 665 -19 -Asaia_bogorensis GSF 665 -20 -Asaia_siamensis GSF 665 -21 -Asaia_astilbes GSF 665 -22 -Asaia_platycodi GSF 665 -23 -Asaia_krungthepensis GSF 665 -24 -Asaia_prunellae GSF 665 -27 -Serratia -DL GSF 665 -28 -Phage-Kit. Kat GSF 665 -29 -Cyanobacterium-RC 610 GSF 665 -30 -Serratia_marcescens-RH GSF 665 -31 -Bacillus_cibi GSF 665 -32 -Pedobacter-BMA GSF 665 -33 -Flavobacterium-KMS GSF 665 -34 -Flavobacterium_hibernum GSF 665 -36 -Flavobacterium_hydatis GSF 665 -39 -Kaistella_koreensis GSF 665 -40 -Kaistella_haifense 217, 320 1, 317, 872 809, 893 301, 171 794, 482 656, 143 595, 655 573, 447 170, 895 593, 179 986, 925 827, 533 297, 153 823, 488 883, 287 1, 098, 516 549, 616 206, 973 1, 096, 204 820, 818 783, 447 808, 325 1, 152, 811 1, 035, 414 129, 258 53, 773 909, 265 307, 886 693, 101 1, 200, 365 185, 975 1, 432, 517 744, 893 1, 238, 892 1, 067, 969 130, 391, 966 790, 723, 170 485, 935, 870 180, 702, 758 476, 689, 384 393, 685, 659 357, 393, 201 344, 068, 354 102, 536, 927 355, 907, 159 592, 154, 880 496, 519, 794 178, 292, 067 494, 092, 592 529, 972, 260 659, 109, 346 329, 769, 324 124, 183, 611 657, 722, 373 492, 490, 968 470, 068, 239 484, 994, 710 691, 686, 698 621, 248, 288 77, 554, 903 32, 263, 632 545, 559, 194 184, 731, 584 415, 860, 714 720, 218, 713 111, 585, 274 859, 510, 422 446, 935, 512 743, 334, 928 640, 781, 490 Total Average 25, 364, 460 724, 699 15, 218, 675, 963 434, 819, 313

Assembly statistics – discussed in Intro and Micro [Soft. Genetics Assembler: Assembly Results Statistics Report] • Total Reads Number: 2056329 • Matched Reads Number: 1983986 • Unmatched Reads Number: 72343 • Assembled Sequences Number: 61 • Average Sequence Length: 57497 • Minimum Sequence Length: 158 • Maximum Sequence Length: 641985 • N 50 Length: 366076 [Final Contig Merge Results Statistics Report] • Final Contig Merge Sequences Number: 13 • Final Contig Merge Average Sequence Length: 269063 • Final Contig Merge Minimum Sequence Length: 173 • Final Contig Merge Maximum Sequence Length: 856388 • Final Contig Merge N 50 Length: 586767 • Matched Reads Count: 1977550 • Number of Matched Bases: 562514128 • Average Read Length: 285 • Average Coverage: 161 • Reference Length: 3507364
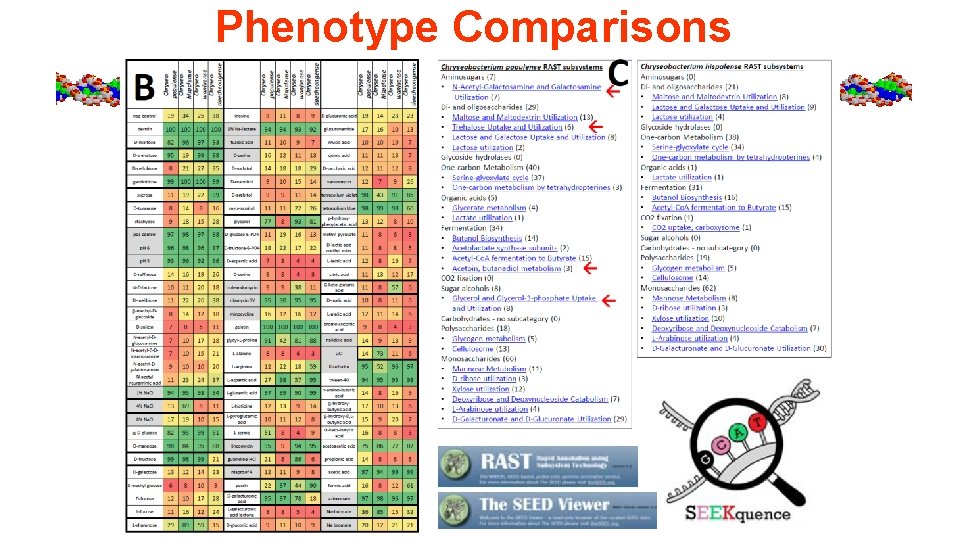
Phenotype Comparisons
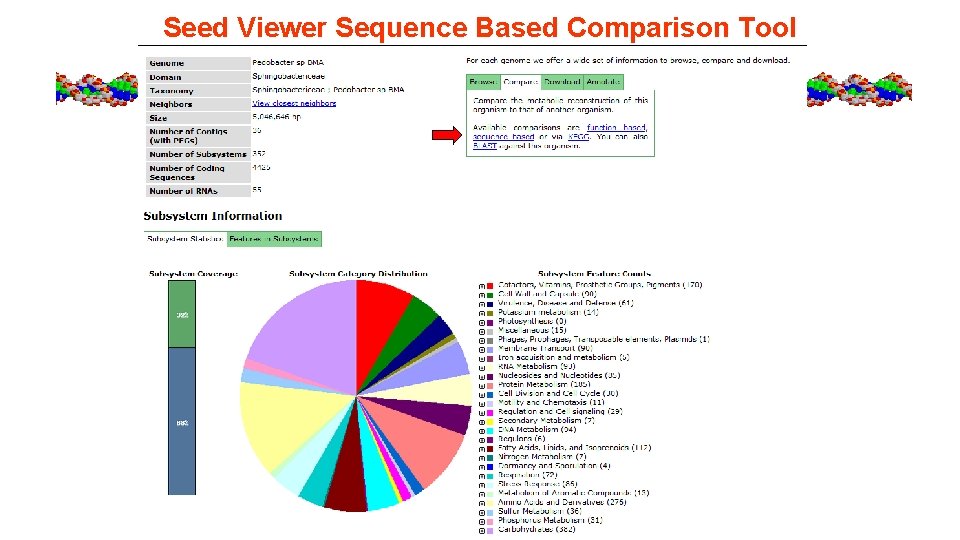
Seed Viewer Sequence Based Comparison Tool
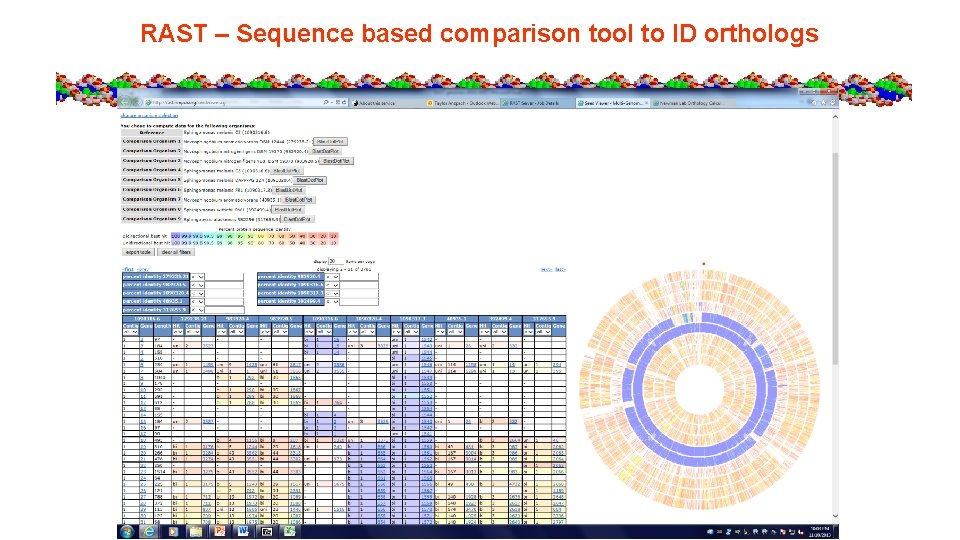
RAST – Sequence based comparison tool to ID orthologs
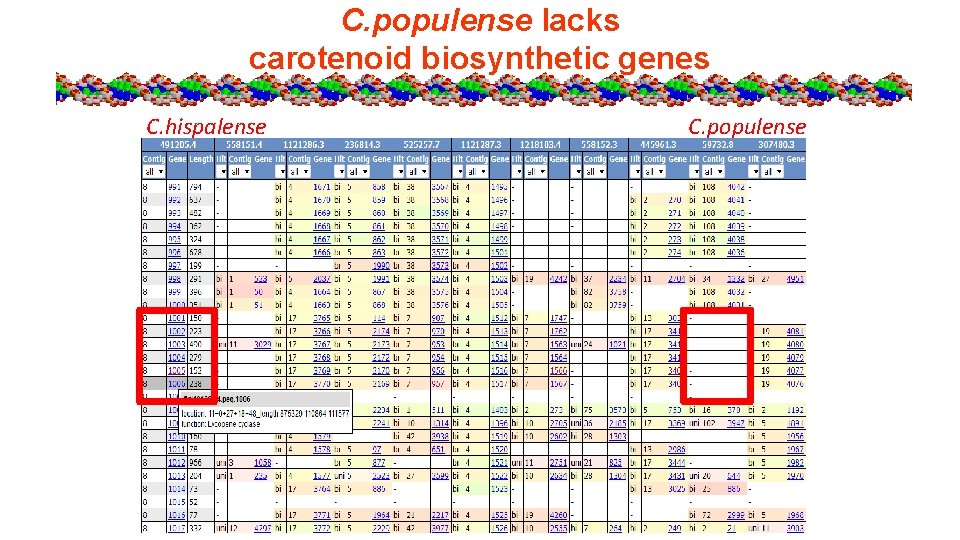
C. populense lacks carotenoid biosynthetic genes C. hispalense C. populense
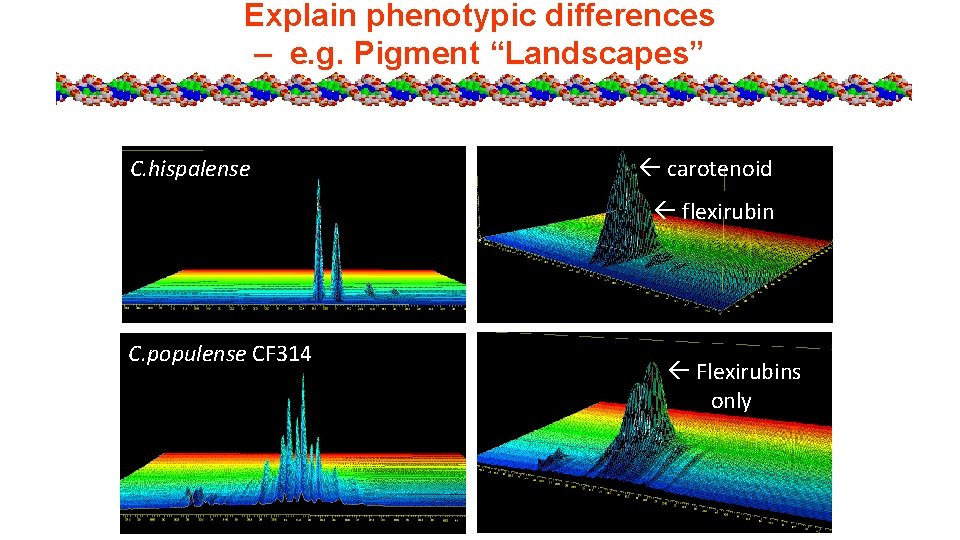
Explain phenotypic differences – e. g. Pigment “Landscapes” C. hispalense carotenoid flexirubin C. populense CF 314 Flexirubins only
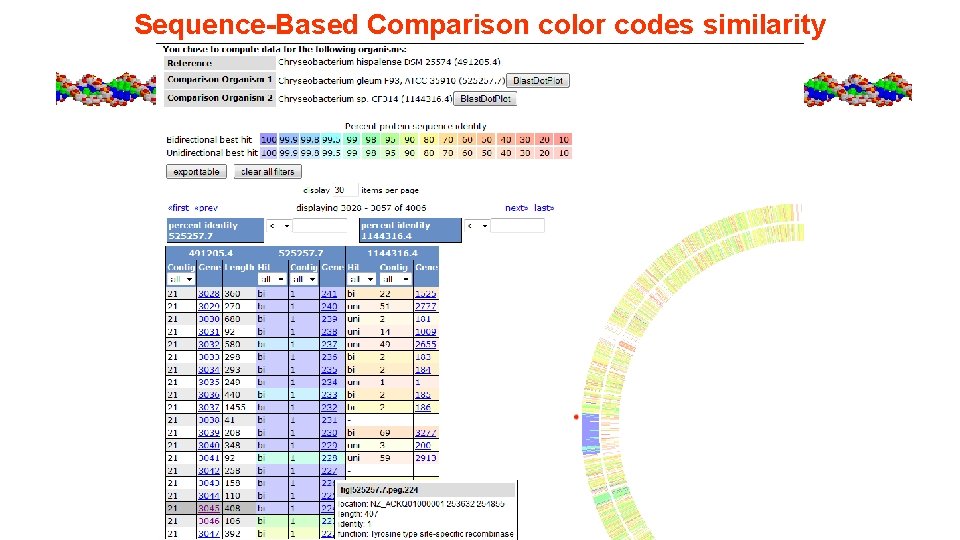
Sequence-Based Comparison color codes similarity
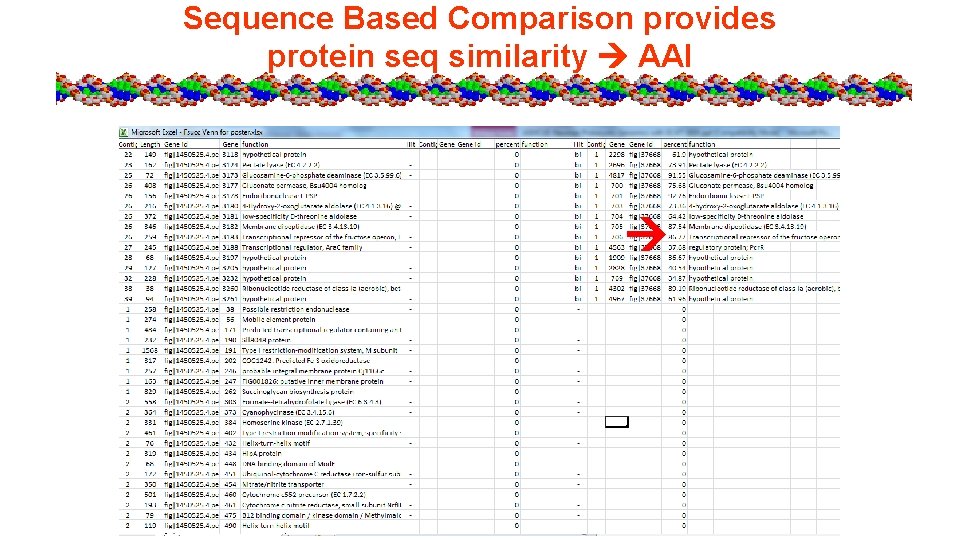
Sequence Based Comparison provides protein seq similarity AAI
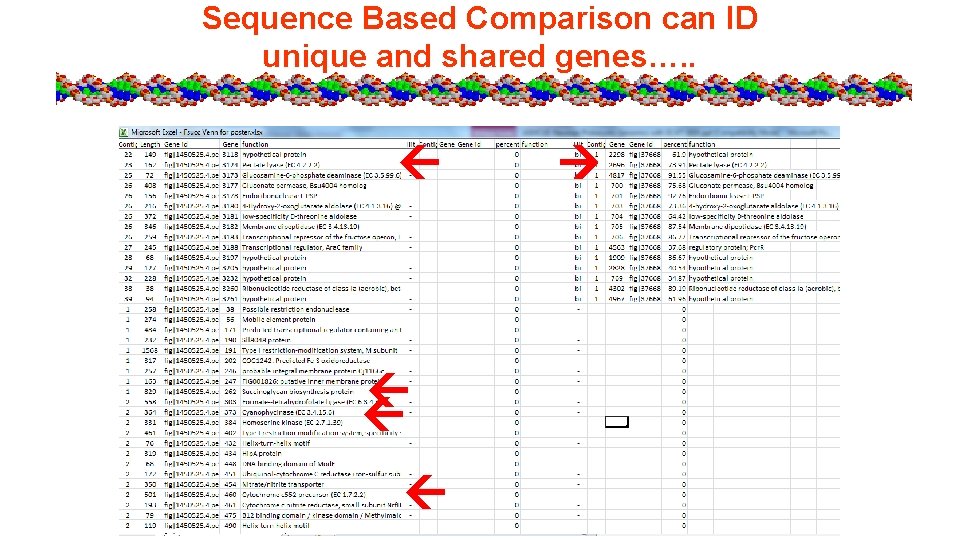
Sequence Based Comparison can ID unique and shared genes…. .
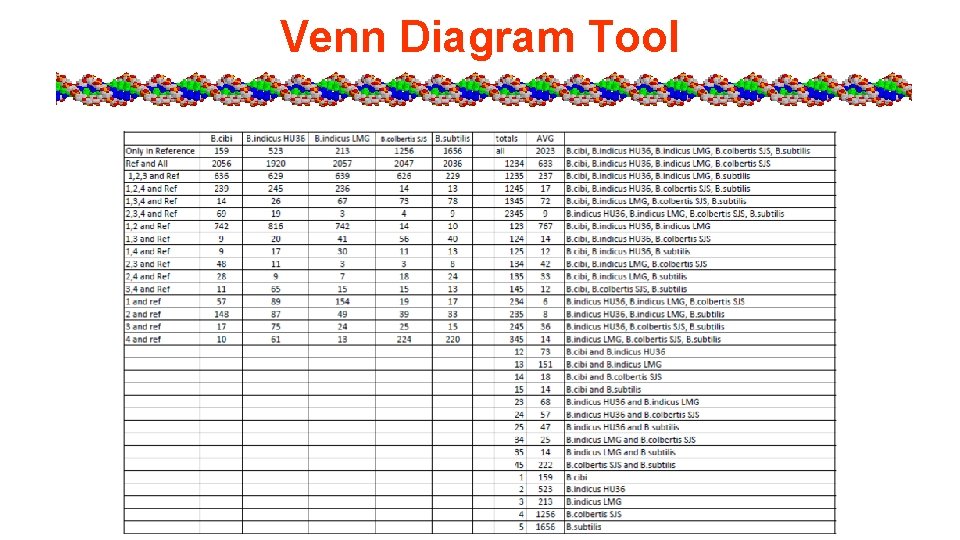
Venn Diagram Tool
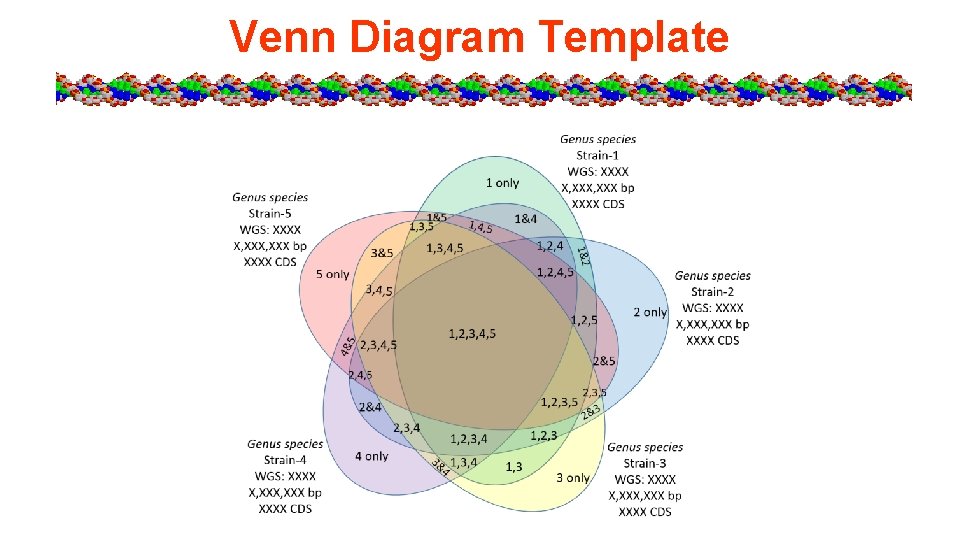
Venn Diagram Template
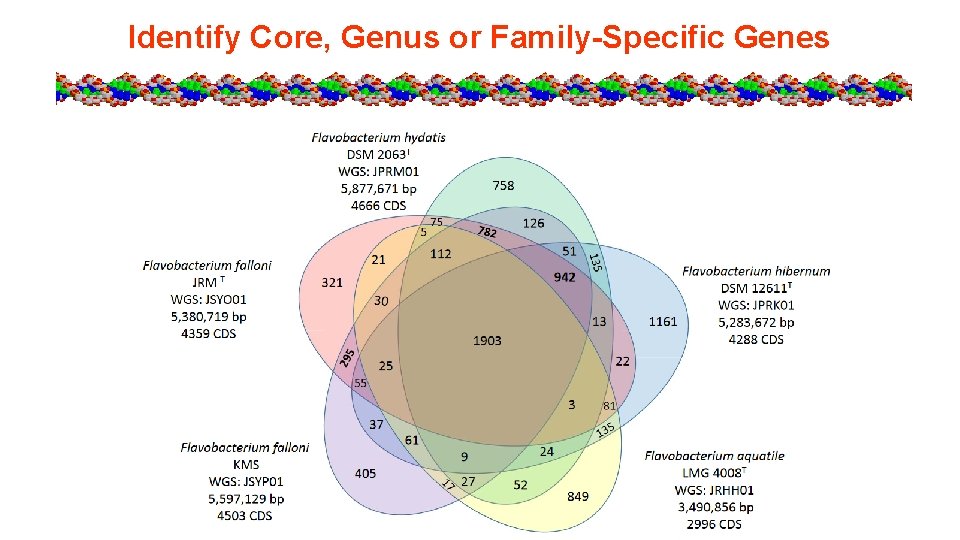
Identify Core, Genus or Family-Specific Genes
Links/Tools available at novelmicrobe. com
- Slides: 49