Developing A Standardsbased Signal Detection and Validation Framework

Developing A Standards-based Signal Detection and Validation Framework of Immune-related Adverse Events Using the OHDSI Common Data Model Yue Yu, Ph. D, Kathryn J. Ruddy, MD, MPH, Shintaro Tsuji, Ph. D, Na Hong, Ph. D, Nilay Shah, Ph. D, Guoqian Jiang, MD, Ph. D Mayo Clinic © 2017 MFMER | slide-1

Background • Immunotherapy has recently been found to be useful in the treatment of certain cancers. • However, immunotherapy for cancer has been associated with unique immune-related adverse events (ir. AEs). Picture Source: https: //www. nobelprize. org/ 2 © 2017 MFMER | slide-2
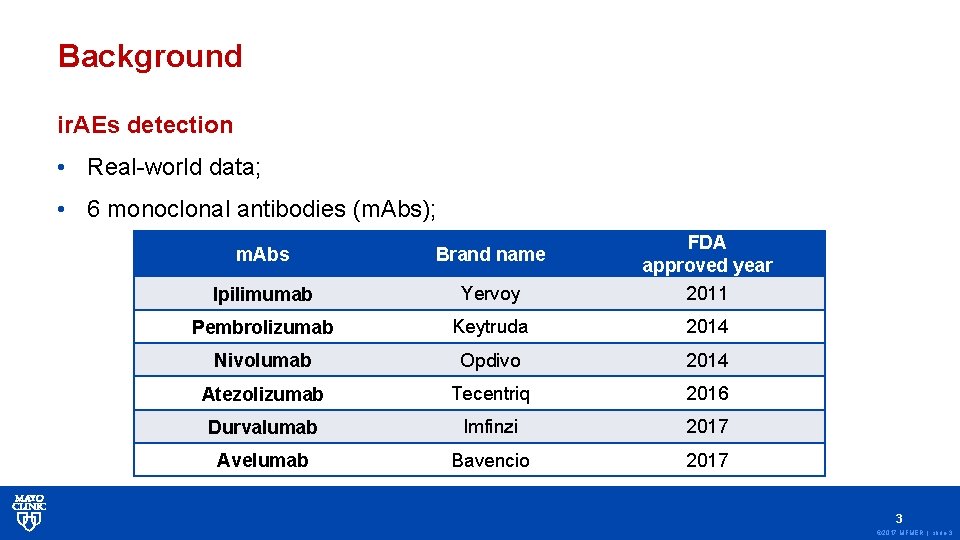
Background ir. AEs detection • Real-world data; • 6 monoclonal antibodies (m. Abs); m. Abs Brand name Ipilimumab Yervoy FDA approved year 2011 Pembrolizumab Keytruda 2014 Nivolumab Opdivo 2014 Atezolizumab Tecentriq 2016 Durvalumab Imfinzi 2017 Avelumab Bavencio 2017 3 © 2017 MFMER | slide-3
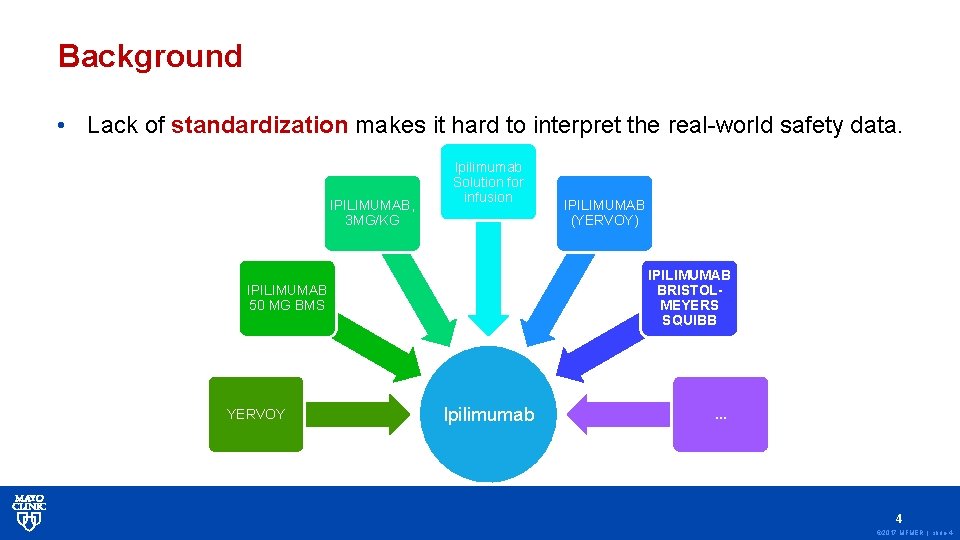
Background • Lack of standardization makes it hard to interpret the real-world safety data. IPILIMUMAB, 3 MG/KG Ipilimumab Solution for infusion IPILIMUMAB BRISTOLMEYERS SQUIBB IPILIMUMAB 50 MG BMS YERVOY IPILIMUMAB (YERVOY) Ipilimumab . . . 4 © 2017 MFMER | slide-4
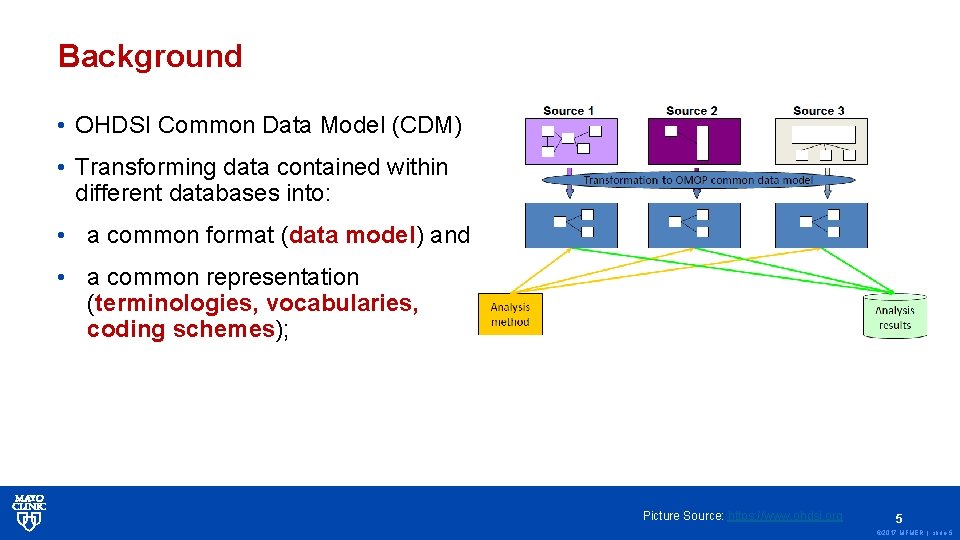
Background • OHDSI Common Data Model (CDM) • Transforming data contained within different databases into: • a common format (data model) and • a common representation (terminologies, vocabularies, coding schemes); Picture Source: https: //www. ohdsi. org 5 © 2017 MFMER | slide-5

Objective • 1. Building a standardized adverse drug reaction detection data platform based on OHDSI CDM; • 2. Developing a comprehensive (real-world data mining + text mining + validation) ir. AEs detection framework. 6 © 2017 MFMER | slide-6

Materials 1) Data standardization & ir. AEs data mining: • FDA Adverse Event Reporting System(FAERS): • • • a database that contains adverse event reports, medication error reports and product quality complaints resulting in adverse events that were submitted to FDA. Widely used in post-marketing safety surveillance by FDA and research communities. ADEpedia-on-OHDSI platform: • • • Try to integrate a spontaneous reporting system data (such as FAERS) and longitudinal observational databases (such as Electronic Health Records, EHRs) An extraction, transformation and loading (ETL) tool to covert FAERS database into the OHDSI CDM format. https: //github. com/adepedia-on-ohdsi 7 © 2017 MFMER | slide-7

Materials • 2) ir. AEs text-mining: • Drug Labels: • 6 m. Abs drug labels in Structured Product Labeling (SPL) format download from Daily. Med. • Publication: • 679 Pub. Med abstracts regarding with m. Abs-ir. AEs. • c. TAKES: • • discovers clinical-named entities and clinical events using a dictionary lookup algorithm and a subset of the UMLS. Common Terminology Criteria for Adverse Events (CTCAE) terminology was used as dictionary to extract ir. AEs concepts from text. 8 © 2017 MFMER | slide-8
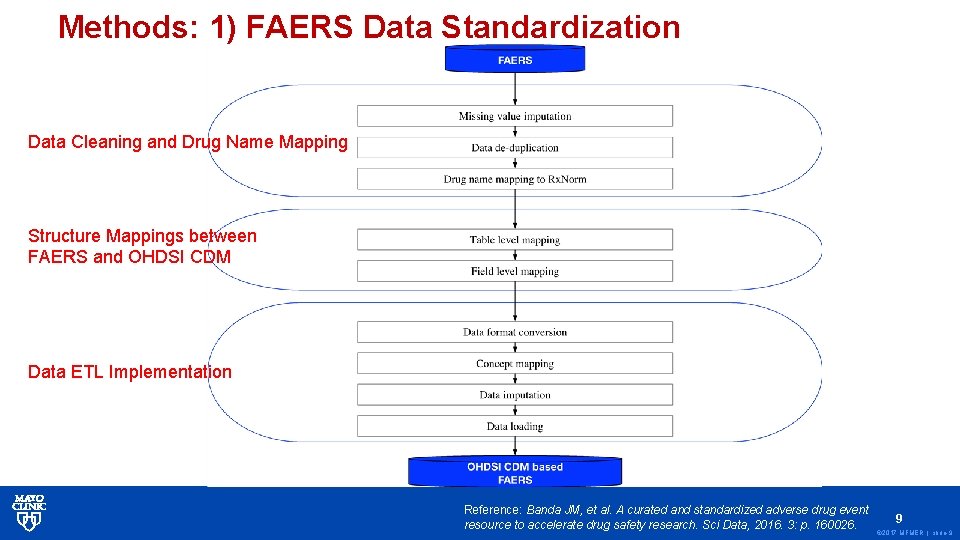
Methods: 1) FAERS Data Standardization Data Cleaning and Drug Name Mapping Structure Mappings between FAERS and OHDSI CDM Data ETL Implementation Reference: Banda JM, et al. A curated and standardized adverse drug event resource to accelerate drug safety research. Sci Data, 2016. 3: p. 160026. 9 © 2017 MFMER | slide-9
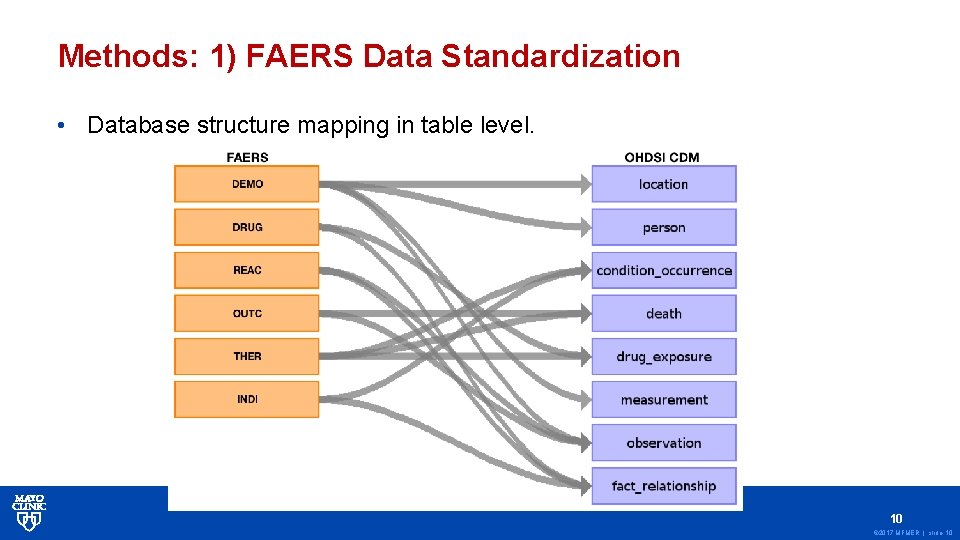
Methods: 1) FAERS Data Standardization • Database structure mapping in table level. 10 © 2017 MFMER | slide-10
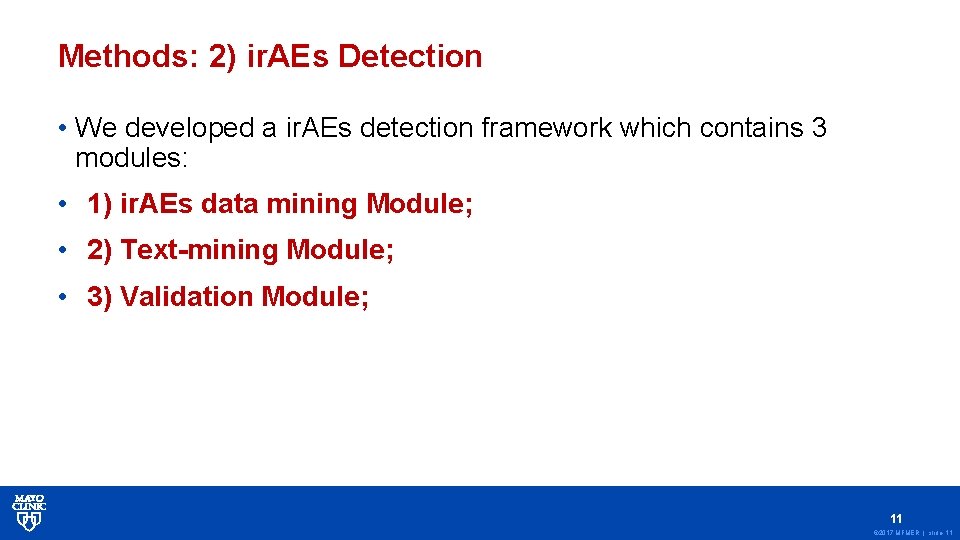
Methods: 2) ir. AEs Detection • We developed a ir. AEs detection framework which contains 3 modules: • 1) ir. AEs data mining Module; • 2) Text-mining Module; • 3) Validation Module; 11 © 2017 MFMER | slide-11
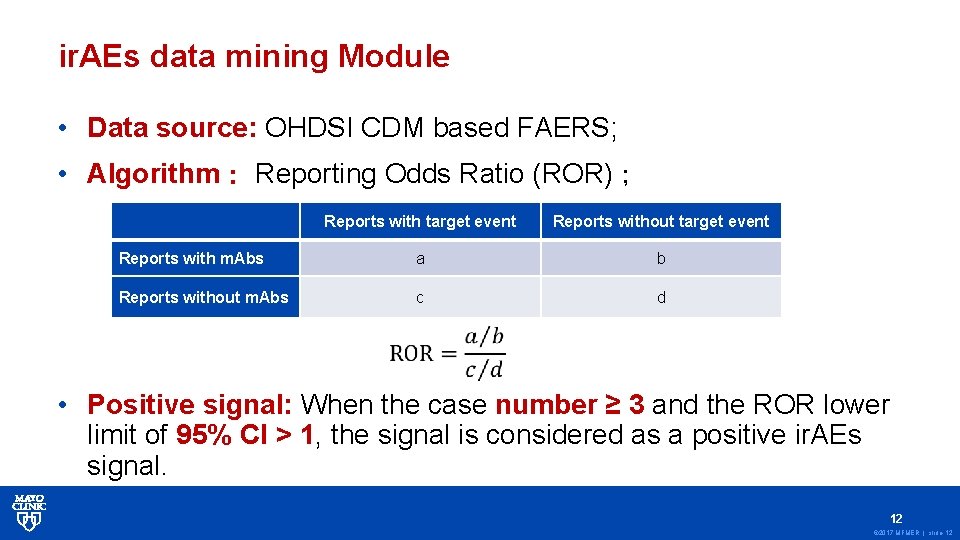
ir. AEs data mining Module • Data source: OHDSI CDM based FAERS; • Algorithm: Reporting Odds Ratio (ROR); Reports with target event Reports without target event Reports with m. Abs a b Reports without m. Abs c d • Positive signal: When the case number ≥ 3 and the ROR lower limit of 95% CI > 1, the signal is considered as a positive ir. AEs signal. 12 © 2017 MFMER | slide-12
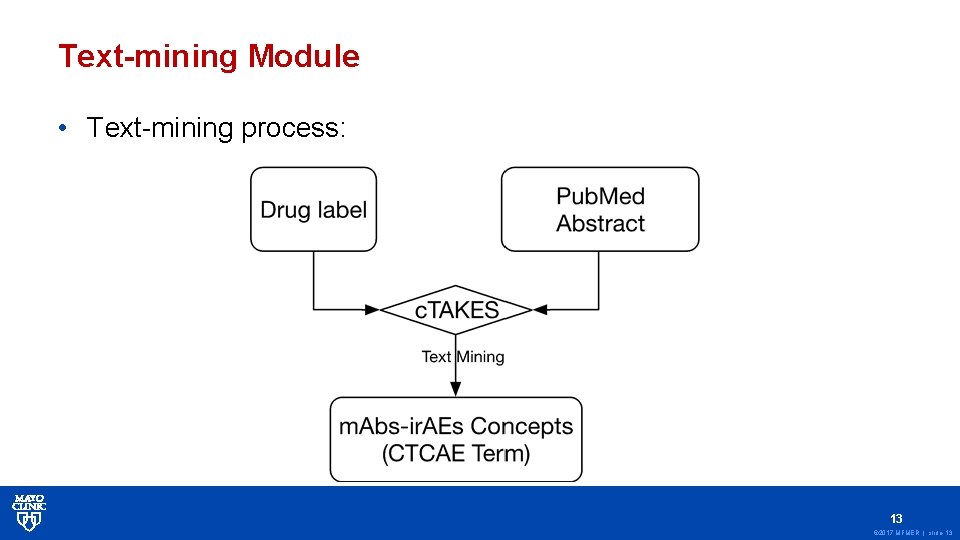
Text-mining Module • Text-mining process: 13 © 2017 MFMER | slide-13
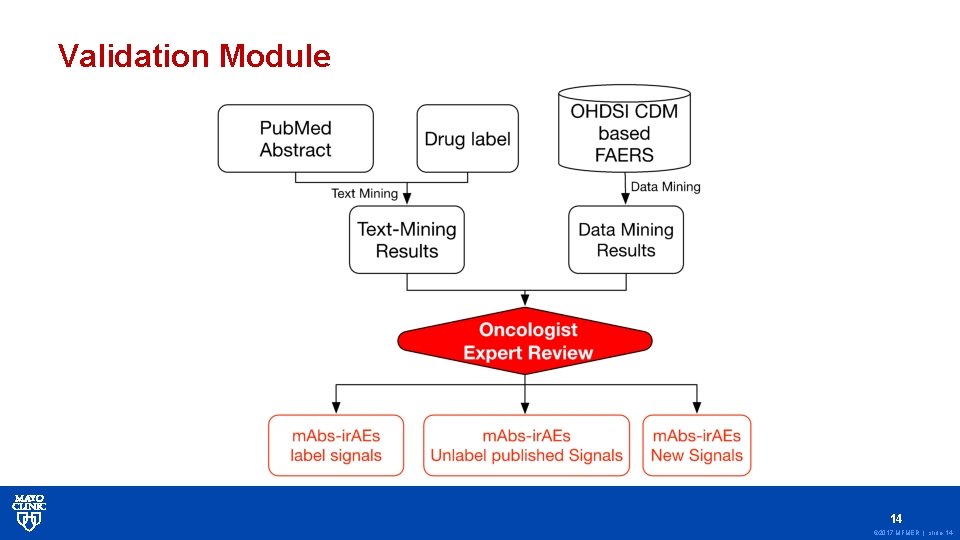
Validation Module 14 © 2017 MFMER | slide-14
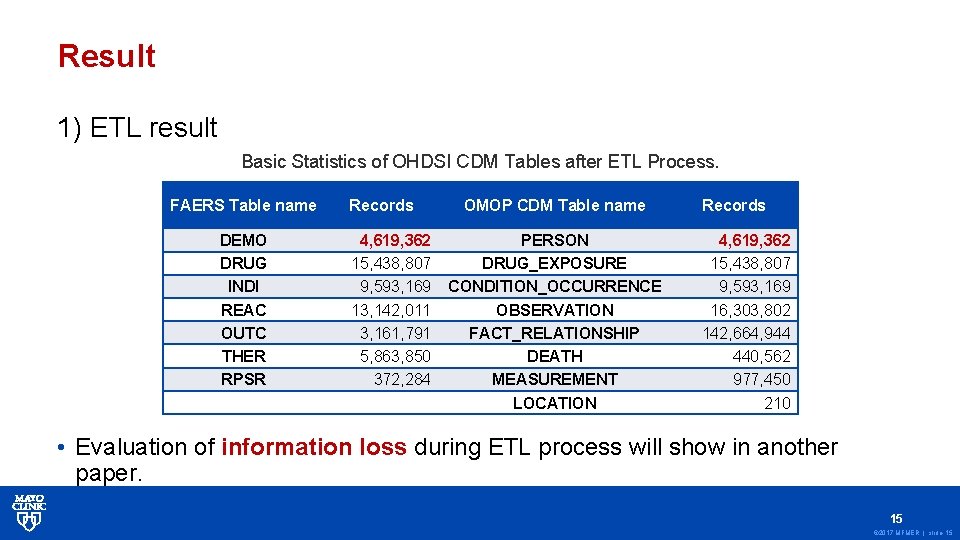
Result 1) ETL result Basic Statistics of OHDSI CDM Tables after ETL Process. FAERS Table name Records DEMO DRUG INDI REAC OUTC THER RPSR 4, 619, 362 15, 438, 807 9, 593, 169 13, 142, 011 3, 161, 791 5, 863, 850 372, 284 OMOP CDM Table name PERSON DRUG_EXPOSURE CONDITION_OCCURRENCE OBSERVATION FACT_RELATIONSHIP DEATH MEASUREMENT LOCATION Records 4, 619, 362 15, 438, 807 9, 593, 169 16, 303, 802 142, 664, 944 440, 562 977, 450 210 • Evaluation of information loss during ETL process will show in another paper. 15 © 2017 MFMER | slide-15
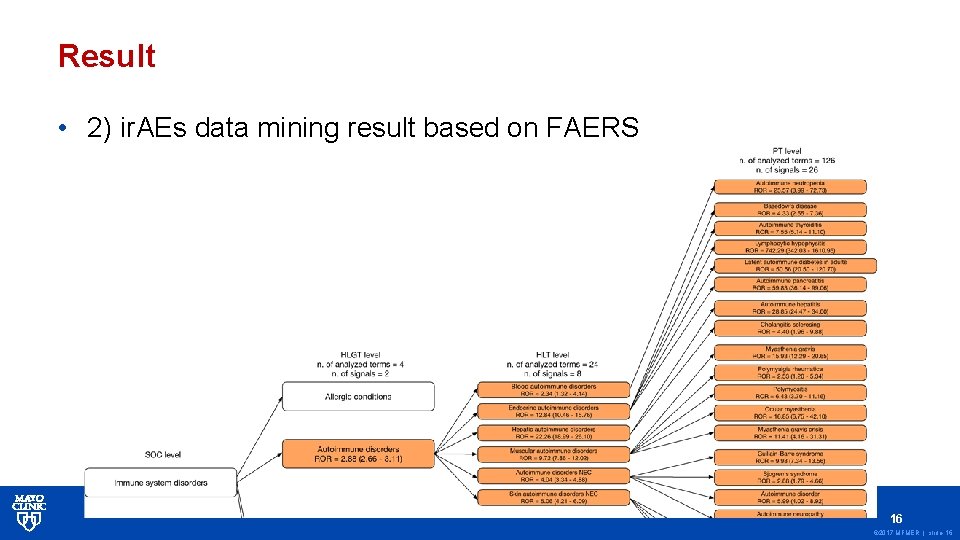
Result • 2) ir. AEs data mining result based on FAERS 16 © 2017 MFMER | slide-16
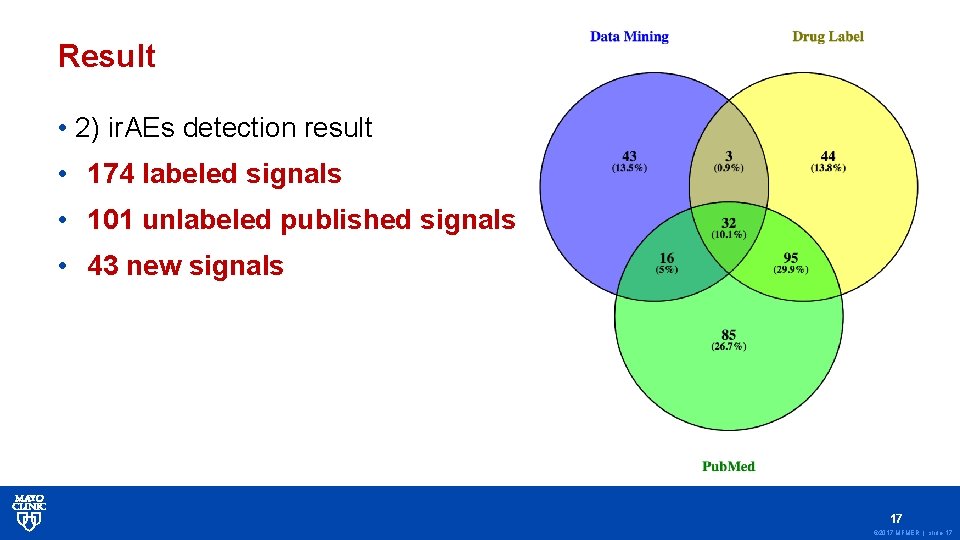
Result • 2) ir. AEs detection result • 174 labeled signals • 101 unlabeled published signals • 43 new signals 17 © 2017 MFMER | slide-17
Discussion & Conclusion Discussion Conclusion • 1) More EHR data and analysis tools • 1) We utilized standard OHDSI will be integrated into our ADEpedia-on CDM to represent the FAERS data. -OHDSI platform in future. • 2) We developed a standards • 2) For the text-mining performance, we based signal detection and conduct the evaluation of CTCAE for validation framework to detect capturing ir. AEs in another paper. ir. AEs signals. • 3) In future study, we will also perform a further human expert review to check whether those newly identified signals are real new signals or just synonyms/subtypes. 18 © 2017 MFMER | slide-18

Thank you! Email me at: Yu. Yue 1@mayo. edu Jiang. Guoqian@mayo. edu
- Slides: 19