Detecting short indels and complex structural variations from

















- Slides: 52

Detecting short indels and complex structural variations from next-gen sequence data Kai Ye Leiden University Medical Center k. ye@lumc. nl

Start Pindel • • Copy Pindel. tar. gz to your local folder tar xvfz Pindel. tar. gz. /run Return to presentation 2

Types of sequencing • • Without a reference genome: New species With a reference genome: Re-sequencing ü common variants in populations SNPs, indels and structural variants ü All (somatic) variants for subjects with extreme phenotypes, such as cancer a) Normal/tumor paired genomes from the same patient b) Exome sequencing of a group of cancer patients

Types of sequencing • • Without a reference genome: New species With a reference genome: Re-sequencing ü common variants in populations SNPs, indels and structural variants ü All (somatic) variants for subjects with extreme phenotypes, such as cancer a) Normal/tumor paired genomes from the same patient b) Exome sequencing of a group of cancer patients

Sequence for common variants in populations Nature, 2010 Nature, 2011

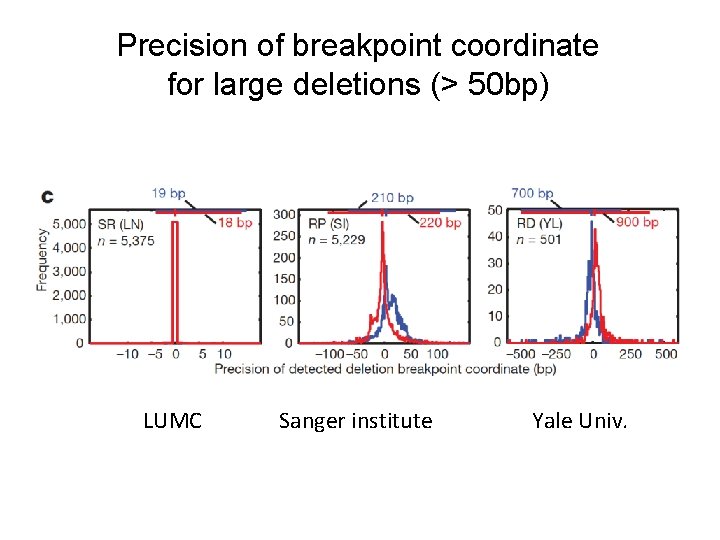
Precision of breakpoint coordinate for large deletions (> 50 bp) LUMC Sanger institute Yale Univ.

Types of sequencing • • Without a reference genome: New species With a reference genome: Re-sequencing ü common variants in populations SNPs, indels and structural variants ü All (somatic) variants for subjects with extreme phenotypes, such as cancer a) Normal/tumor paired genomes from the same patient b) Exome sequencing of a group of cancer patients

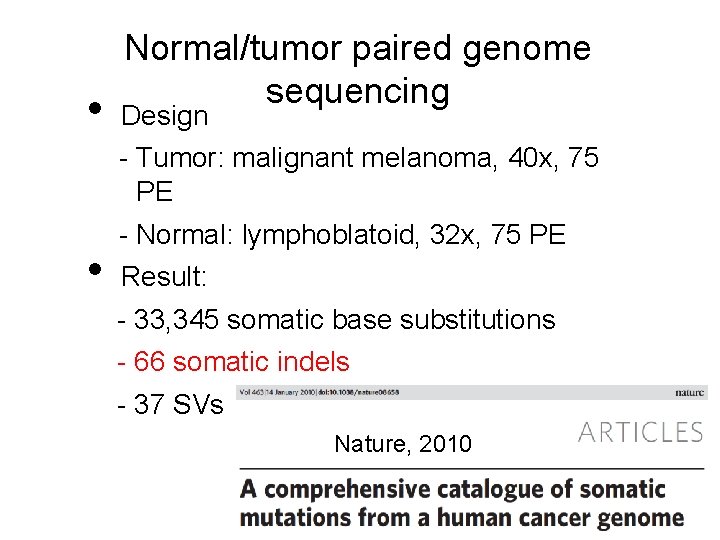
• Normal/tumor paired genome sequencing Design - Tumor: malignant melanoma, 40 x, 75 PE • - Normal: lymphoblatoid, 32 x, 75 PE Result: - 33, 345 somatic base substitutions - 66 somatic indels - 37 SVs Nature, 2010 8 8

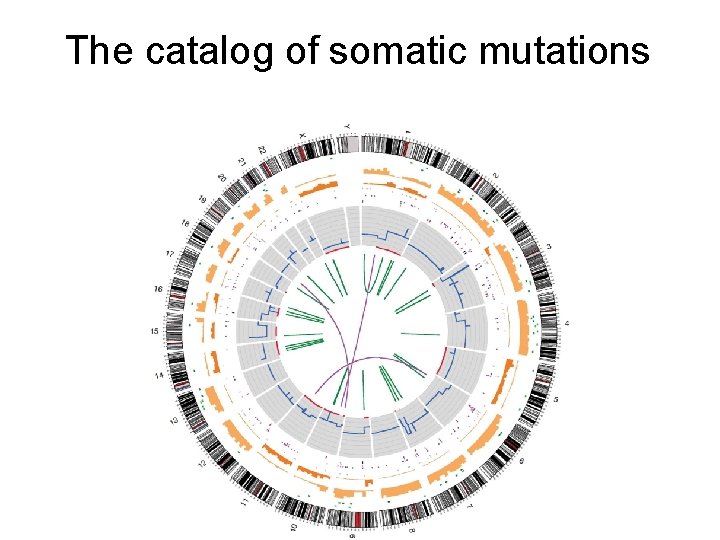
The catalog of somatic mutations

Types of sequencing • • Without a reference genome: New species With a reference genome: Re-sequencing ü common variants in populations SNPs, indels and structural variants ü All (somatic) variants for subjects with extreme phenotypes, such as cancer a) Normal/tumor paired genomes from the same patient b) Exome sequencing of a group of cancer patients

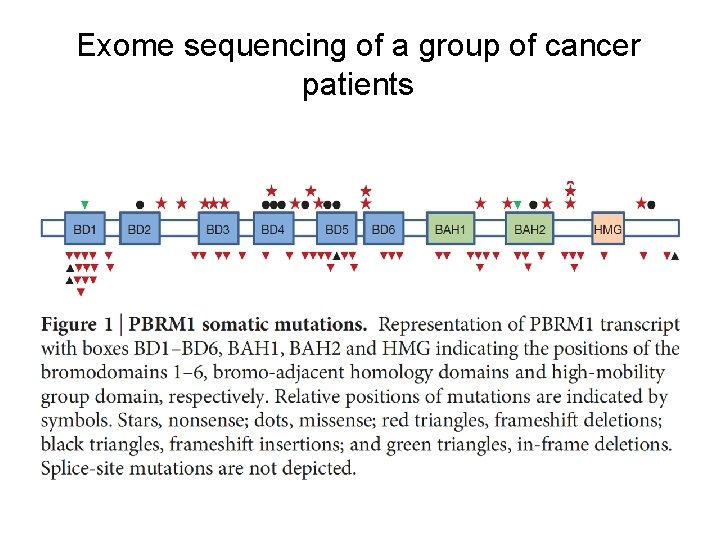
Exome sequencing of a group of cancer patients • • PBRM 1 with truncating mutations in 41% (92/227) of cases. My tool, Pindel, was used to detect somatic indels. Nature, 2011 11

Exome sequencing of a group of cancer patients

From sequence data to variants samples Sequencing, processing data and variant calling Biological discovery

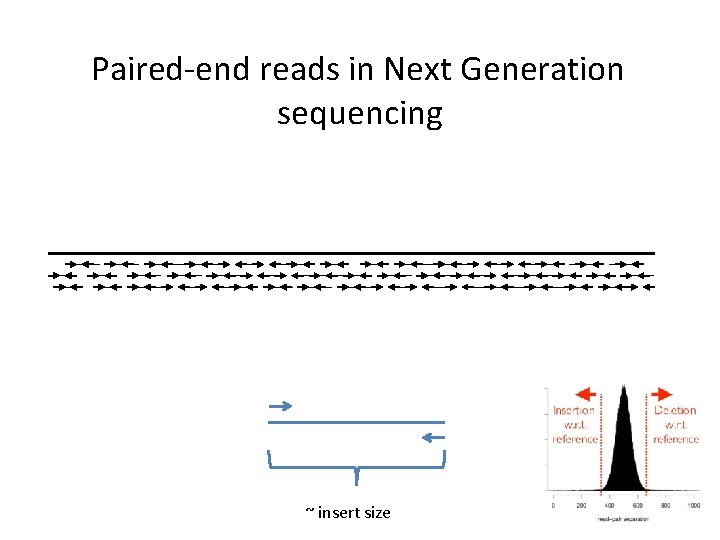
Paired-end reads in Next Generation sequencing

Paired-end reads in Next Generation sequencing ~ insert size

Substitution ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCAGCAGATCAGTC CAGT

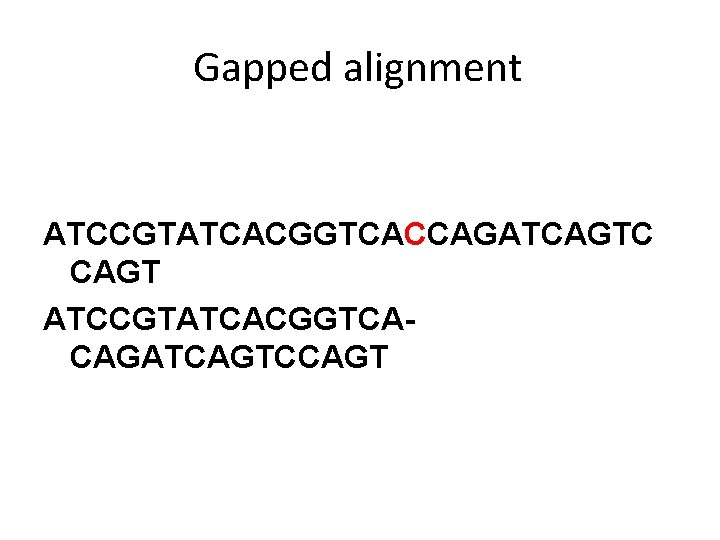
Gapped alignment ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCACAGATCAGTCCAGT

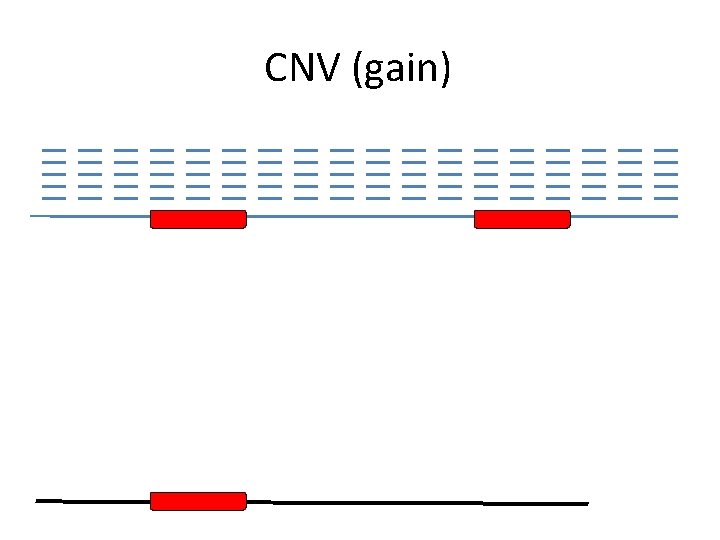
CNV (gain)

CNV (loss)

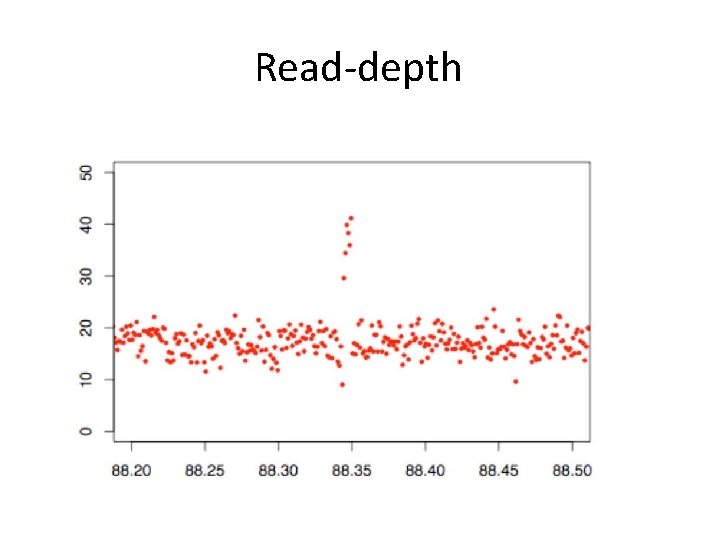
Read-depth

Read-pair approach Sample No Indel Reference Sample Deletion Reference Sample Insertion Reference

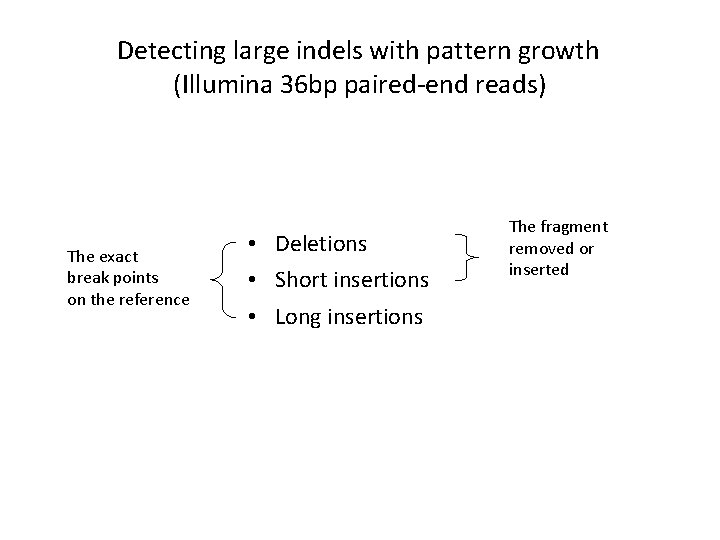
Detecting large indels with pattern growth (Illumina 36 bp paired-end reads) The exact break points on the reference • Deletions • Short insertions • Long insertions The fragment removed or inserted

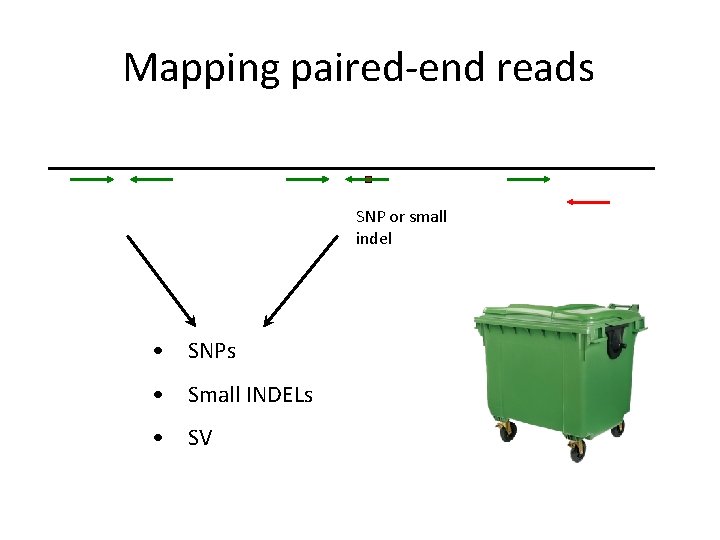
Mapping paired-end reads SNP or small indel • SNPs • Small INDELs • SV

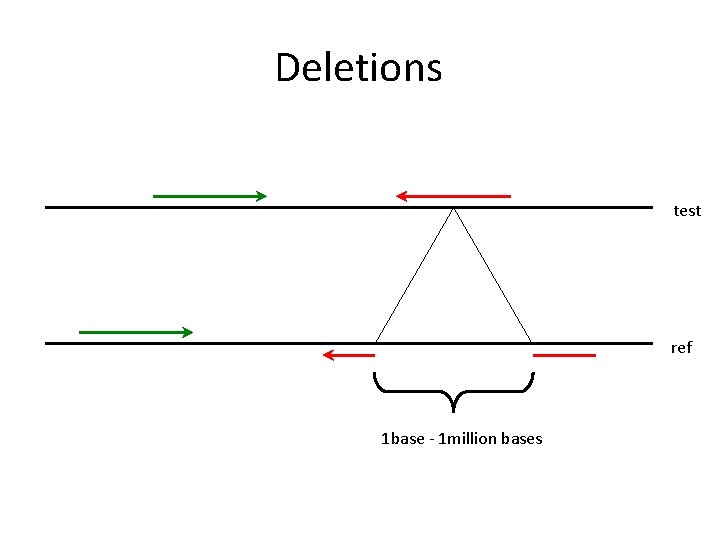
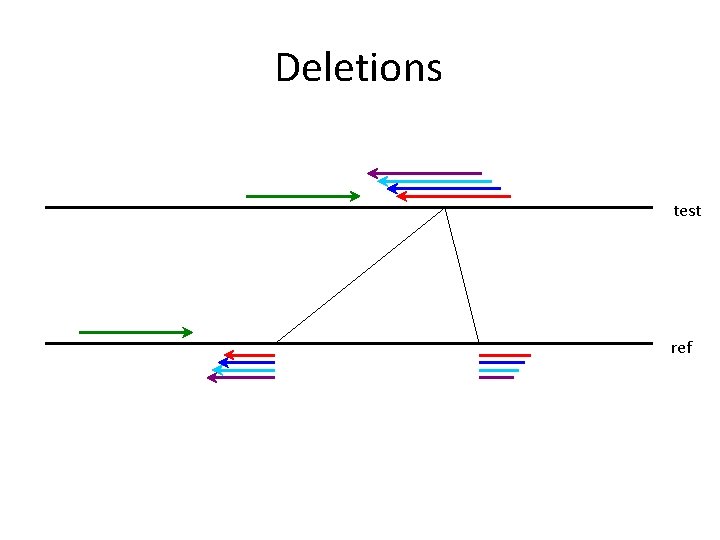
Deletions test ref 1 base - 1 million bases

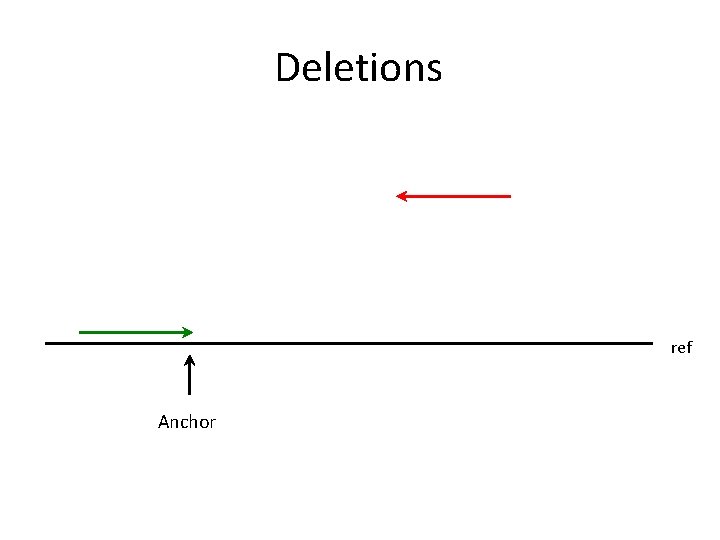
Deletions ref Anchor

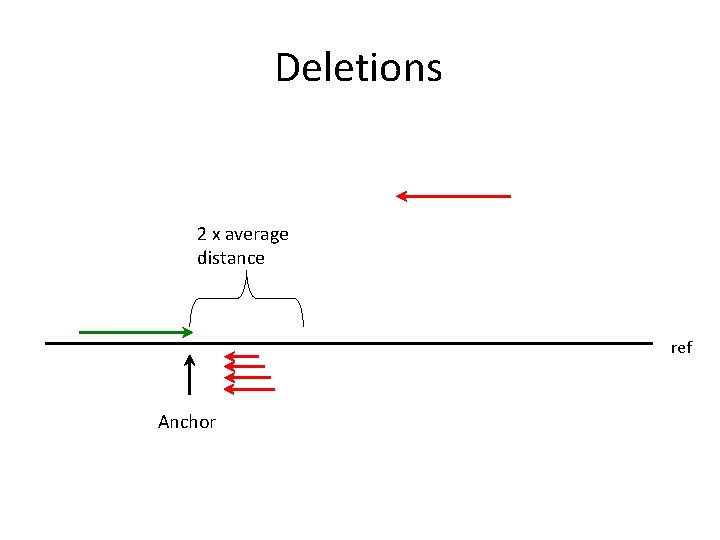
Deletions 2 x average distance ref Anchor

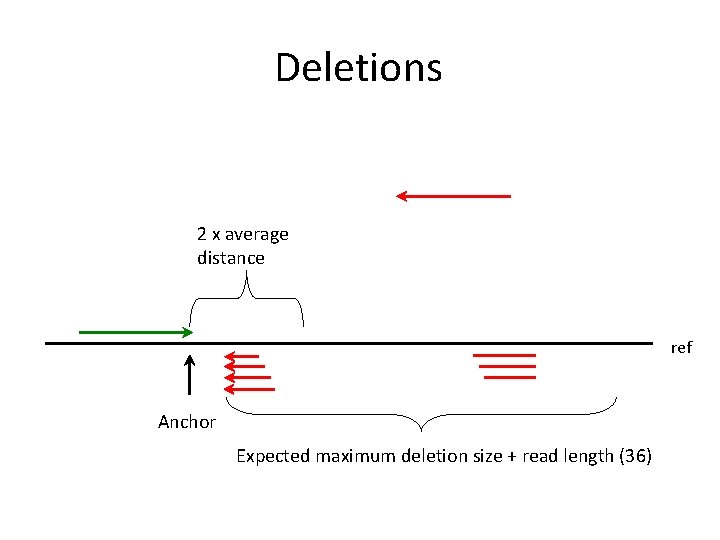
Deletions 2 x average distance ref Anchor Expected maximum deletion size + read length (36)

Deletions test ref

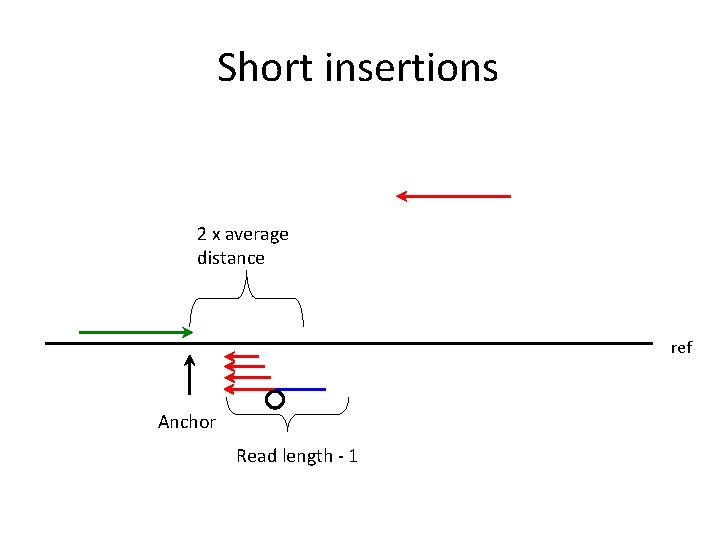
Short insertions 2 x average distance ref Anchor Read length - 1

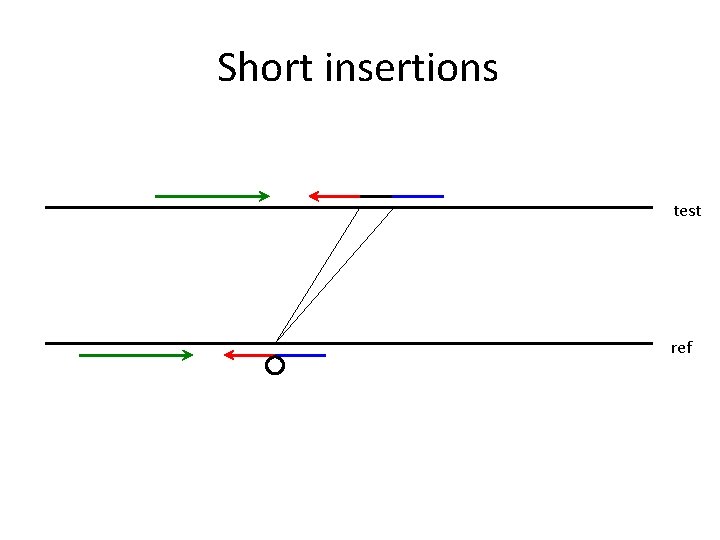
Short insertions test ref

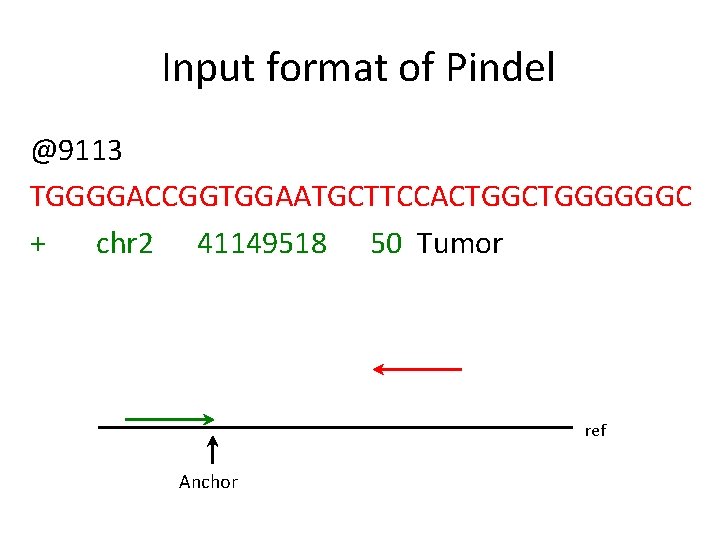
Input format of Pindel @9113 TGGGGACCGGTGGAATGCTTCCACTGGGGGGC + chr 2 41149518 50 Tumor ref Anchor

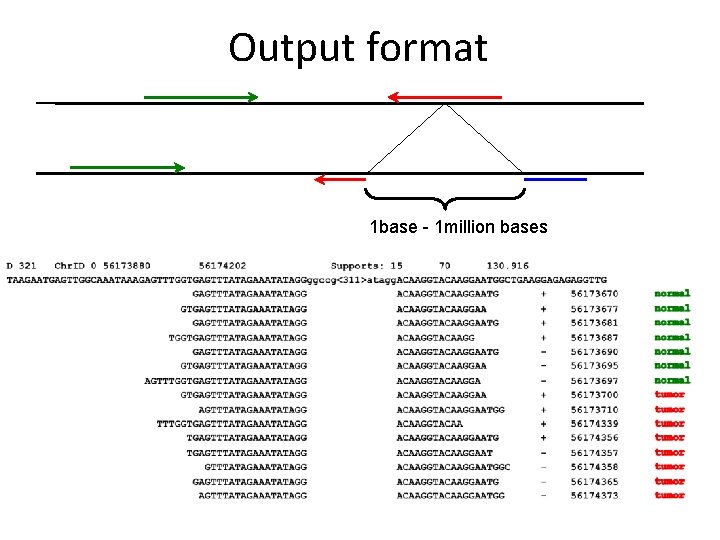
Output format 1 base - 1 million bases

Breakpoint shift ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCAGCAGATCAGTC CAGT ATCCGTATCACGGAAATGCTGTCAGTC CAGT ATCCGTATCACGGAATGCTGTCAGTCCAGT ATCCGTATCACGGATATGCAAGCAGTC CAGT ATCCGTATCACGG--

Breakpoint shift ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCAGCAGATCAGTC CAGT ATCCGTATCACGGAAATGCTGTCAGTC CAGT ATCCGTATCACGGAATGCTGTCAGTCCAGT ATCCGTATCACGGATATGCAAGCAGTC CAGT ATCCGTATCACGG--

Breakpoint shift ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCAGCAGATCAGTC CAGT ATCCGTATCACGGAAATGCTGTCAGTC CAGT ATCCGTATCACGGAATGCTGTCAGTCCAGT ATCCGTATCACGGATATGCAAGCAGTC CAGT ATCCGTATCACGG--

Breakpoint shift ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCAGCAGATCAGTC CAGT ATCCGTATCACGGAAATGCTGTCAGTC CAGT ATCCGTATCACGGAATGCTGTCAGTCCAGT ATCCGTATCACGGATATGCAAGCAGTC CAGT ATCCGTATCACGG--

Breakpoint shift ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCAGCAGATCAGTC CAGT ATCCGTATCACGGAAATGCTGTCAGTC CAGT ATCCGTATCACGGAATGCTGTCAGTCCAGT ATCCGTATCACGGATATGCAAGCAGTC CAGT ATCCGTATCACGGA--

Breakpoint shift ATCCGTATCACGGTCACCAGATCAGTC CAGT ATCCGTATCACGGTCAGCAGATCAGTC CAGT ATCCGTATCACGGAAATGCTGTCAGTC CAGT ATCCGTATCACGGAATGCTGTCAGTCCAGT ATCCGTATCACGGATATGCAAGCAGTC CAGT ATCCGTATCACGGAT--

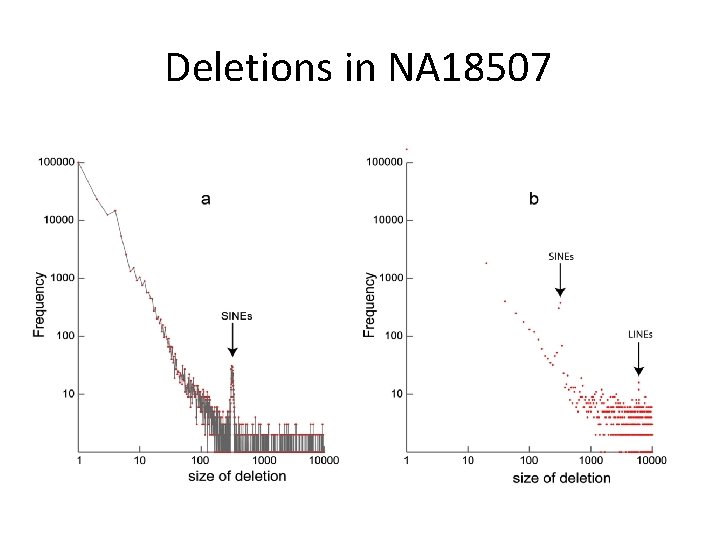
Real data on NA 18507 • Input: – Bentley et al. , 2008 – 2 x 36 bp paired-end reads – 135 Gb of sequence – ~4 billion paired 35 -base reads • Preprocessing: – ~56 M pairs of one-end mapped reads • Output: – 4. 5 hours on a single CPU and maximum memory usage 1. 5 G – 146, 843 deletions (1 bp-10 kb) and 142, 908 short insertions (1 -16 bp) – 91. 2% of 1 -16 bp deletions and 87. 2% of 1 -16 bp insertions overlap with calls from Illumina

Deletions in NA 18507

Paired (cancer) genomes • • COLO-829 cells Normal ~30 x paired-end 75 nt reads Tumor ~40 x paired-end 75 nt reads Search for somatic (tumor specific) indels

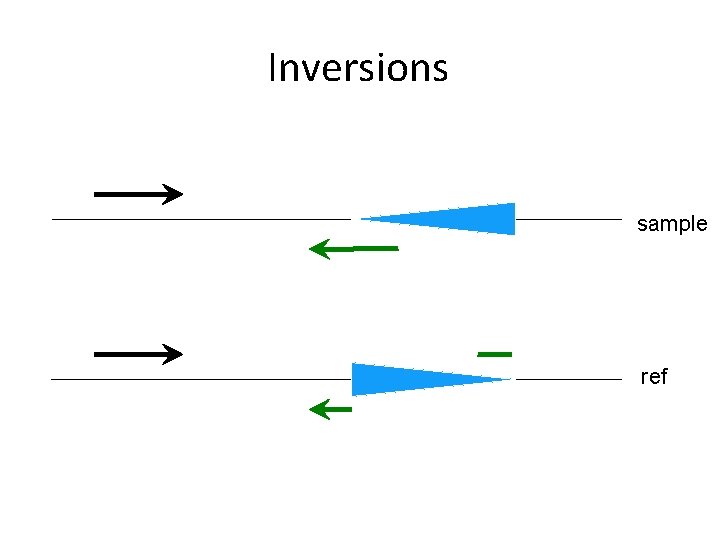
Inversions sample ref

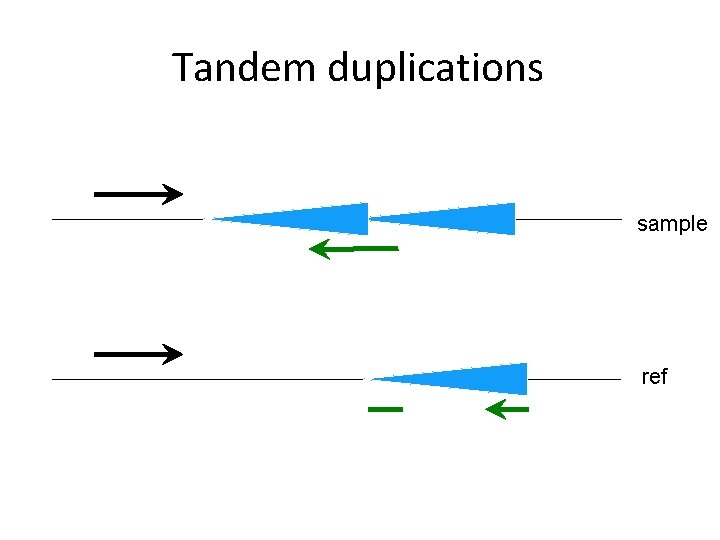
Tandem duplications sample ref

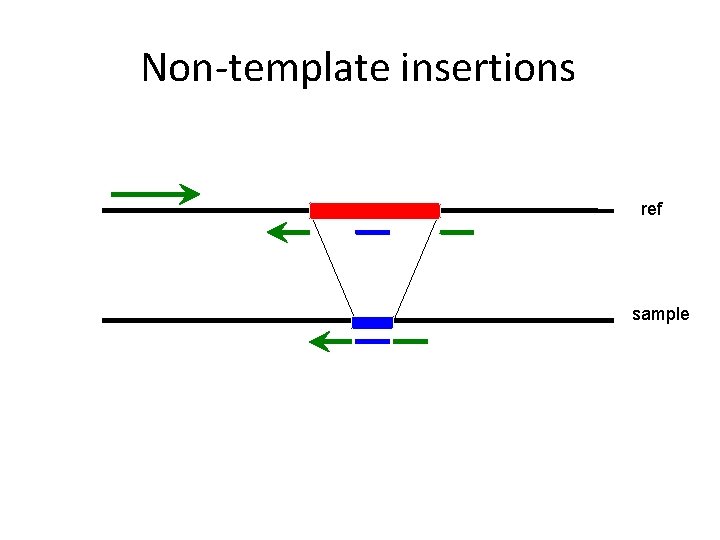
Non-template insertions ref sample

Non-template insertions D 4 I 2 Chr. ID 3 BP 156978978 156978983 Supports 12 + 0 - 12 S 1 13 SUM_MS 627 Num. Sup. Samples 1 HCC 1599 a 12 CATGGCTGACTTATAAATCCCTACAGATATGTGGTTACTTCTCTACTTTCCCTTTGGCTTGGGCAACTGCCACGTTGATGCACTGGAGCCATTCTTCTGCATTCTTCTCATCCTTGGCCTTAA AGACATAGGTTTTATTGTC TTATAAATCCCTACAGATATGTGGTTACTTCTCTACTTTCCCTTTGCCTTGGGCAACTGCCAAA GATGCACT ATGTGGTTACTTCTCTACTTTCCCTTTGGCTTGGGCAACTGCCAAA GATGCACTGGAGCCATTCTTCTGCAT CTCTACTTTCCCTTTGGCTTGGGCAACTGCCA AA GATGCACTGGAGCCATTCTTCTGCATTCTTCTCATCCT AGATATGTGGTTACTTCTCTACTTTCCCTTTGGCTTGGGCAACTGCCAAA GATGCACTGGAGCCATTCTTCT TTTCCCTTTGGCTTGGGCAACTGCCA AA GATGCACTGGAGCCATTCTTCTGCATTCTTCTCATCCTTGGCCTT TTACTTCTCTACTTTCCCTTTGGCTTGGGCAACTGCCA AA GATGCACTGGAGCCATTCTTCTGCATTCTTCTCATCCTTGGCCTTAAAGACATAGGTTT CTACAGATATGTGGTTACTTCTCTACTTTCCCTTTGGCTTGGGCAACTGCCAAA GATGCACTGGAGCCATTC AAATCCCTACAGATATGTGGTTACTTCTCTACTTTCCCTTTGGCTTGGGCAACTGCCAAA GATGCACTGGAG CTTGGGCAACTGCCA AA GATGCACTGGAGCCATTCTTCTGCATTCTTCTCATCCTTGGCCTTAAAGACATAGGTTT TTCCCTTTGGCTTGGGCAACTGCCA AA GATGCACTGGAGCCATTCTTCTGCATTCTTCTCATCCTTGGCCTT

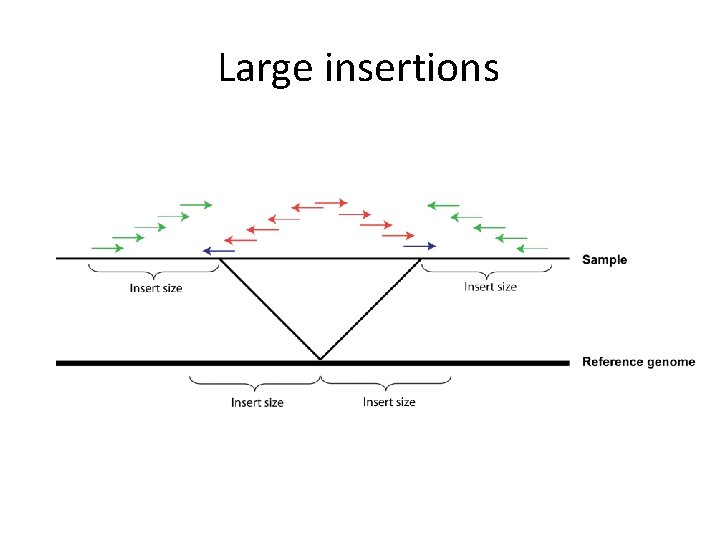
Large insertions

Allow mismatches D 10 Chr. ID 13 BP 32913041 32913052 AAATCAACTAGTGACCTTCCAGGGACAACCCGAACGTGATGAAAAGATCAaagaacctac. TCTATTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTTGGACAAAGT GATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTTGGACAA CAACCCGAACGTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTTGGA TGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTTGGACAAAG GTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTTGGAC TAGTGACCTTCCAGGGACAACCCGAACGTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAA ACAACCCGAACGTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTT CCCGAACGTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATC AACCCGAACGTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTTGGACA ACCTTCCAGGGACAACCCGAACGTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTTGGACAA AACCCGAACGTGATGAAAAGATCA TCTGTTGGGTTTTCATACAGCTAGCGGGAAAAAAGTTAAAATTGCAAAGGAATCTTT

Variant types • • • Short indels Large deletions Tandem duplications Inversions Breakpoints of large insertions - With/without non-template sequence

Where to get Pindel • www. ebi. ac. uk/~kye/pindel • https: //trac. nbic. nl/pindel • ftp. sanger. ac. uk/pub/zn 1/pindel • K. ye@lumc. nl • kye@ebi. ac. uk

WWW. EBI. AC. UK/~KYE/PINDEL K. YE@LUMC. NL KYE@EBI. AC. UK

Practical • chr 20 -p, MZ twin data of Dorret • Qs: – How many reads in file twin_reads. txt – Open output file for deletions • less twin_output/twin_reads. txt_20_D – Do you see micro-satellite and homopolymer events? – Can you find potentially discordant events? https: //trac. nbic. nl/pindel/

• grep Chr. ID twin_output/twin_reads. txt_20_D | awk '{if ($31 == 1 && $16 >= 10) print}'