Details of Hydrophobic Hydration From MD Simulations Tanya

Details of Hydrophobic Hydration From MD Simulations Tanya Raschke Michael Levitt Department of Structural Biology Stanford University
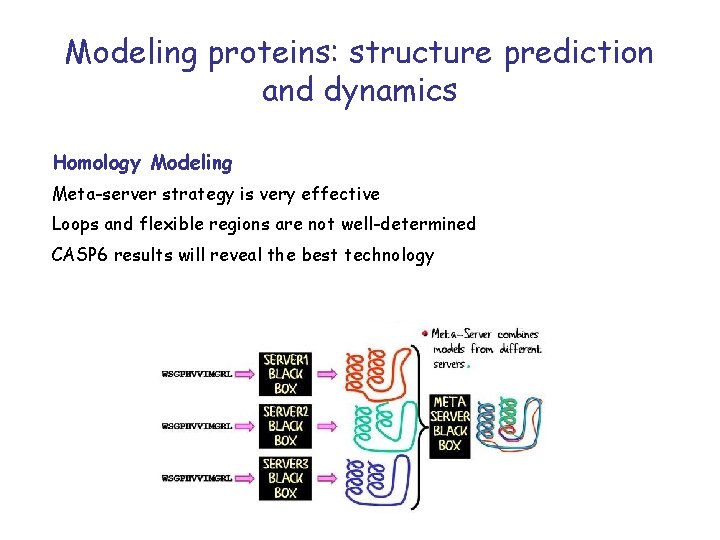
Modeling proteins: structure prediction and dynamics Homology Modeling Meta-server strategy is very effective Loops and flexible regions are not well-determined CASP 6 results will reveal the best technology
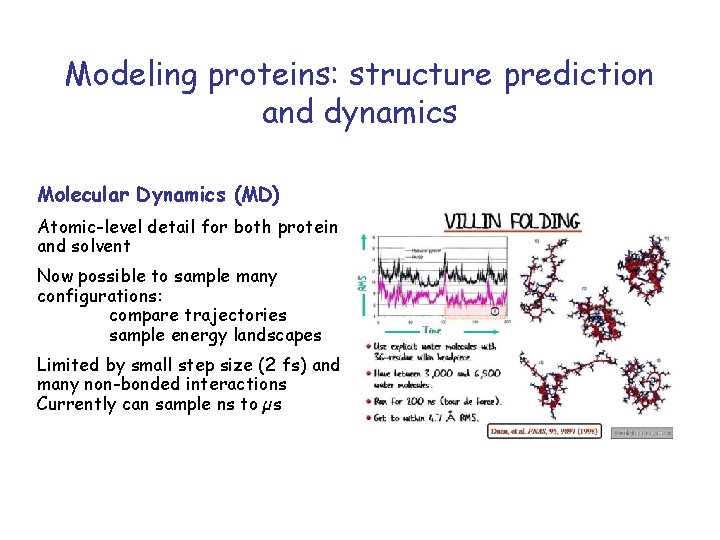
Modeling proteins: structure prediction and dynamics Molecular Dynamics (MD) Atomic-level detail for both protein and solvent Now possible to sample many configurations: compare trajectories sample energy landscapes Limited by small step size (2 fs) and many non-bonded interactions Currently can sample ns to µs
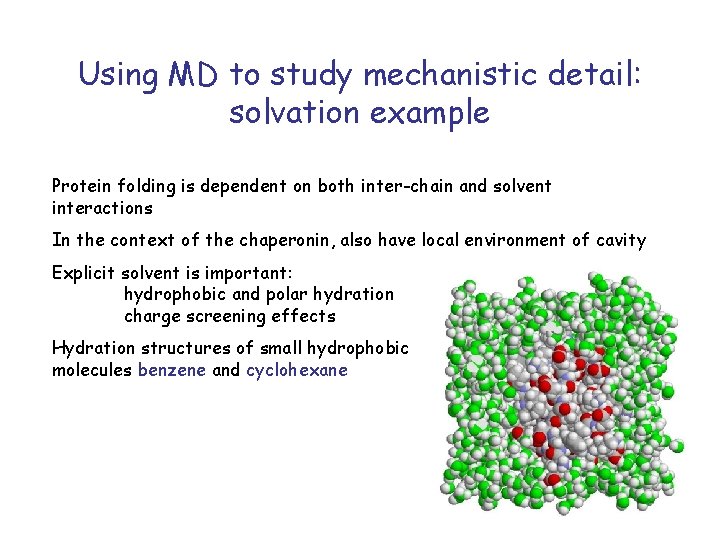
Using MD to study mechanistic detail: solvation example Protein folding is dependent on both inter-chain and solvent interactions In the context of the chaperonin, also have local environment of cavity Explicit solvent is important: hydrophobic and polar hydration charge screening effects Hydration structures of small hydrophobic molecules benzene and cyclohexane
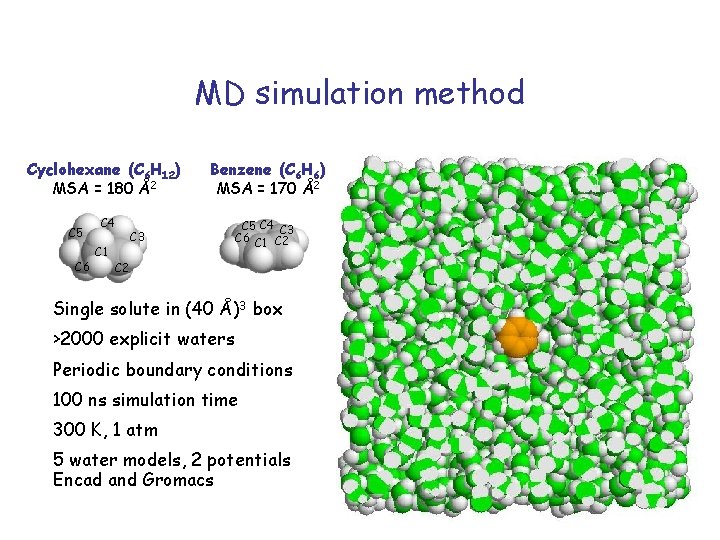
MD simulation method Cyclohexane (C 6 H 12) MSA = 180 Å2 C 5 C 6 C 4 C 1 Benzene (C 6 H 6) MSA = 170 Å2 C 3 C 5 C 4 C 3 C 6 C 1 C 2 Single solute in (40 Å)3 box >2000 explicit waters Periodic boundary conditions 100 ns simulation time 300 K, 1 atm 5 water models, 2 potentials Encad and Gromacs
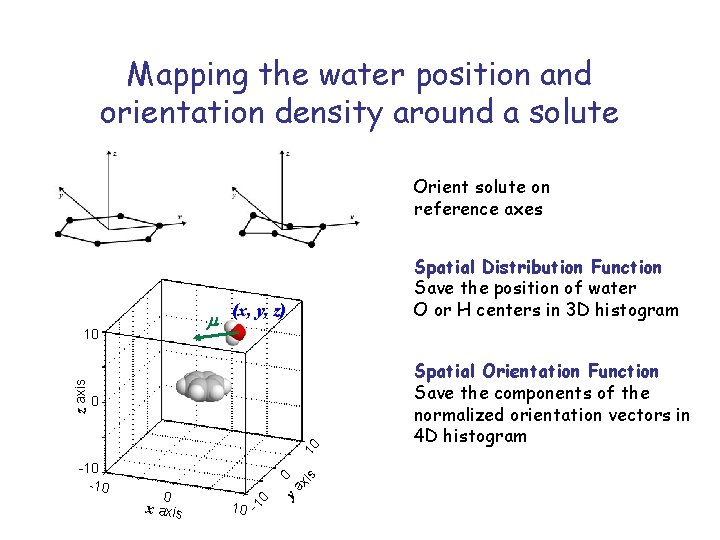
Mapping the water position and orientation density around a solute Orient solute on reference axes m (x, y, z) 0 xi ya 0 0 x axis -1 0 -10 s 10 z axis 10 Spatial Distribution Function Save the position of water O or H centers in 3 D histogram 10 Spatial Orientation Function Save the components of the normalized orientation vectors in 4 D histogram
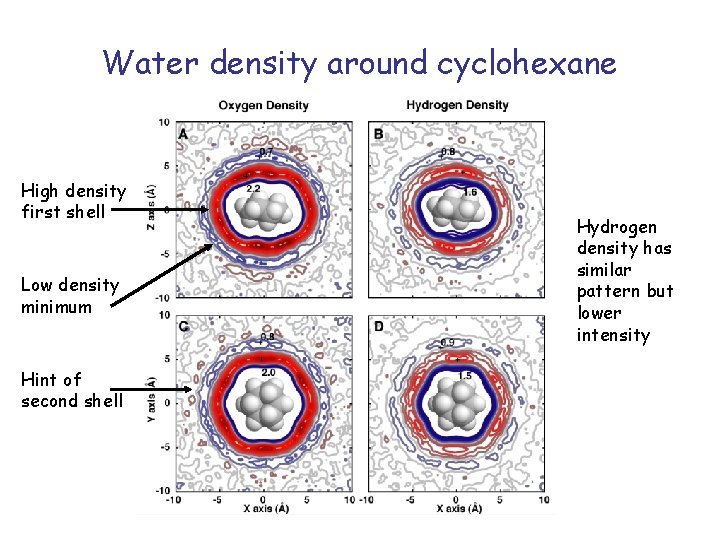
Water density around cyclohexane High density first shell Low density minimum Hint of second shell Hydrogen density has similar pattern but lower intensity
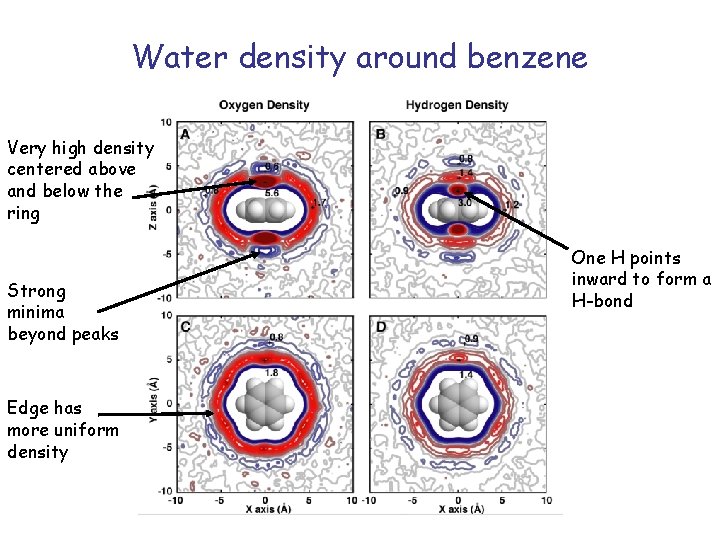
Water density around benzene Very high density centered above and below the ring Strong minima beyond peaks Edge has more uniform density One H points inward to form a H-bond
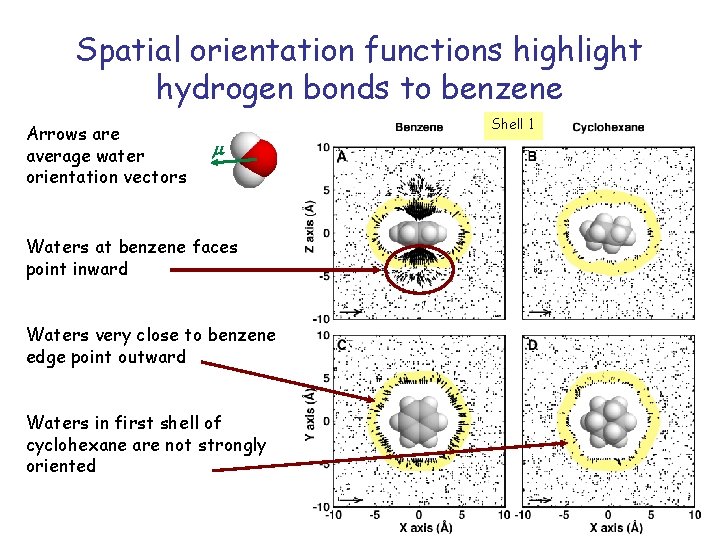
Spatial orientation functions highlight hydrogen bonds to benzene Arrows are average water orientation vectors Shell 1 m Waters at benzene faces point inward Waters very close to benzene edge point outward Waters in first shell of cyclohexane are not strongly oriented
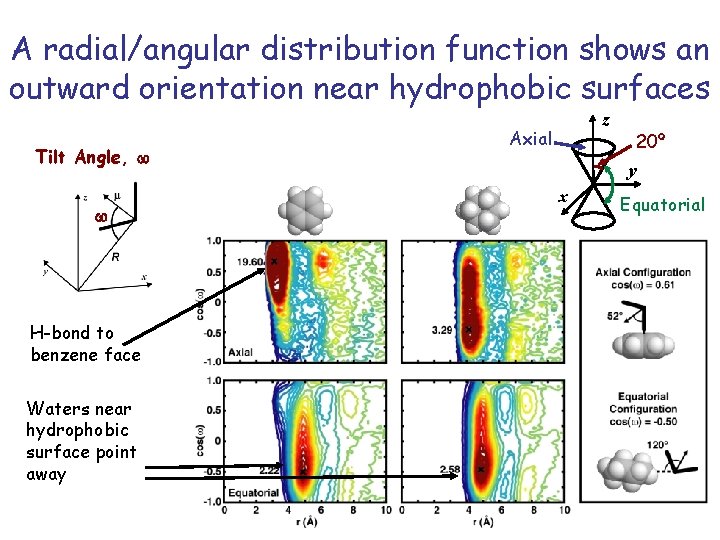
A radial/angular distribution function shows an outward orientation near hydrophobic surfaces Tilt Angle, w w H-bond to benzene face Waters near hydrophobic surface point away z Axial 20º y x Equatorial
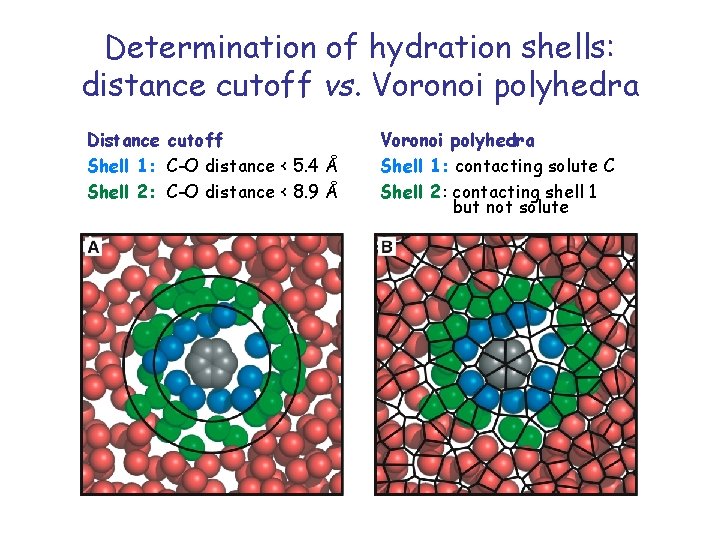
Determination of hydration shells: distance cutoff vs. Voronoi polyhedra Distance cutoff Shell 1: C-O distance < 5. 4 Å Shell 2: C-O distance < 8. 9 Å Voronoi polyhedra Shell 1: contacting solute C Shell 2: contacting shell 1 but not solute
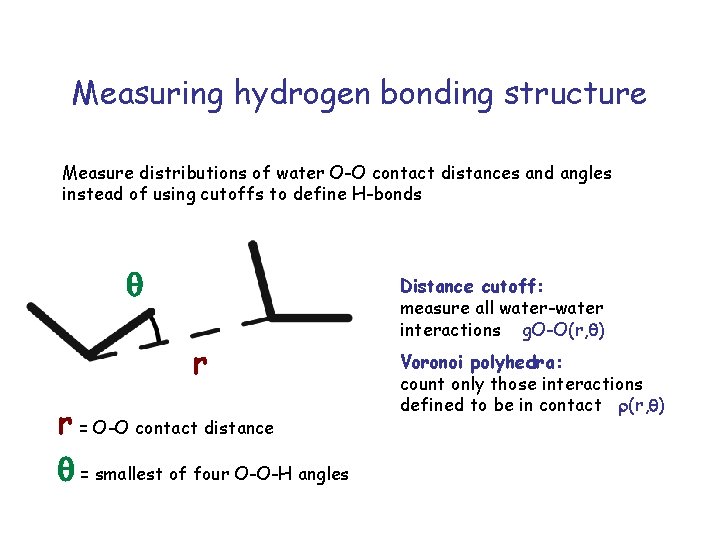
Measuring hydrogen bonding structure Measure distributions of water O-O contact distances and angles instead of using cutoffs to define H-bonds q r r = O-O contact distance q = smallest of four O-O-H angles Distance cutoff: measure all water-water interactions g. O-O(r, q) Voronoi polyhedra: count only those interactions defined to be in contact r(r, q)
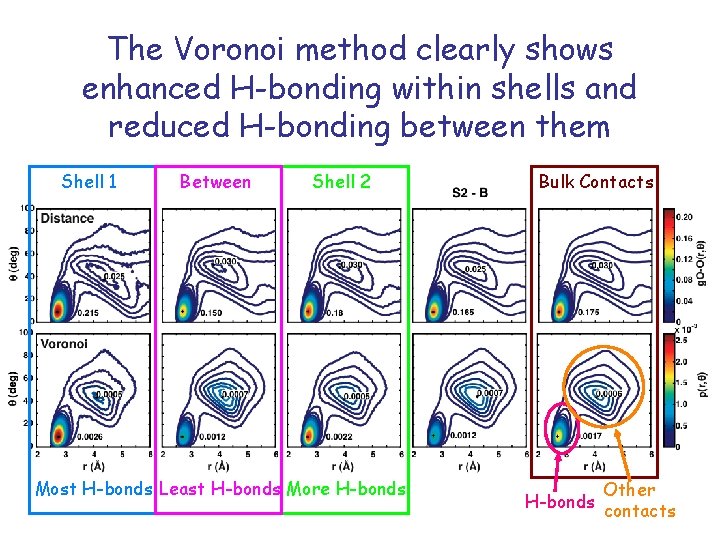
The Voronoi method clearly shows enhanced H-bonding within shells and reduced H-bonding between them Shell 1 Between Shell 2 Most H-bonds Least H-bonds More H-bonds Bulk Contacts H-bonds Other contacts

Summary of hydrophobic hydration Classical explicit solvent models capture the hydrophobic effect Observe preferential H-bonding to benzene faces very high density strong absolute orientation Ordering in first hydration shell of cyclohexane is more subtle high density tangential orientation allows H-bonds but maintains rotational degrees of freedom Hydrogen bonding is significantly enhanced in shell 1 yet markedly reduced between shells 1 and 2
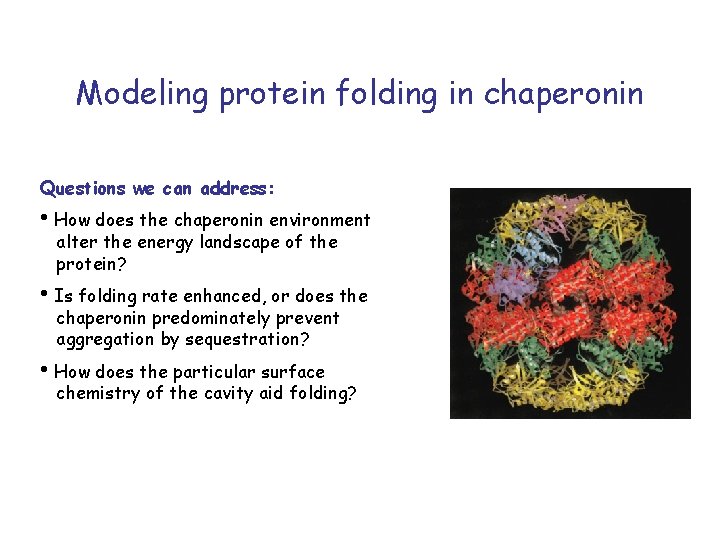
Modeling protein folding in chaperonin Questions we can address: • How does the chaperonin environment alter the energy landscape of the protein? • Is folding rate enhanced, or does the chaperonin predominately prevent aggregation by sequestration? • How does the particular surface chemistry of the cavity aid folding?
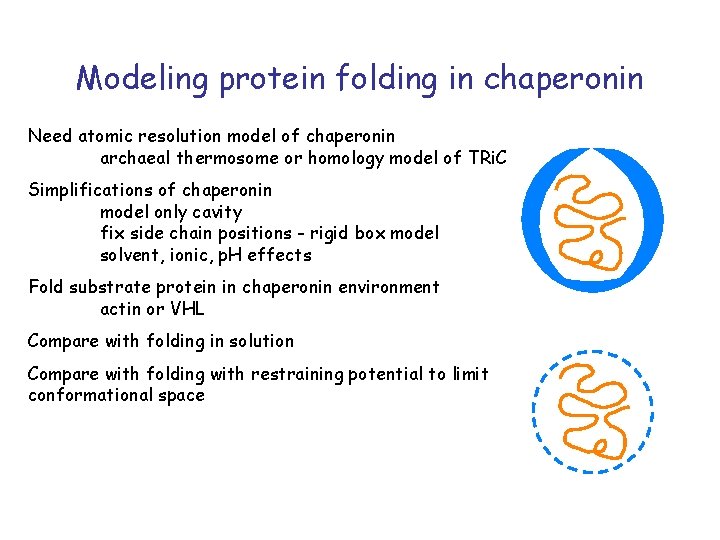
Modeling protein folding in chaperonin Need atomic resolution model of chaperonin archaeal thermosome or homology model of TRi. C Simplifications of chaperonin model only cavity fix side chain positions - rigid box model solvent, ionic, p. H effects Fold substrate protein in chaperonin environment actin or VHL Compare with folding in solution Compare with folding with restraining potential to limit conformational space
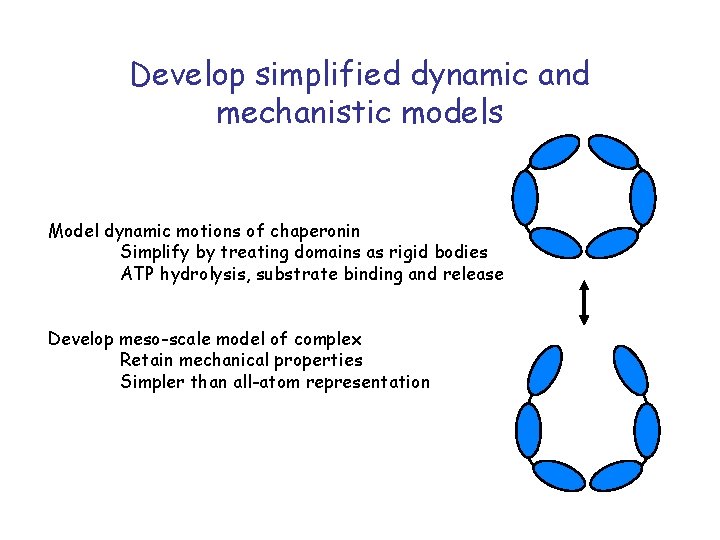
Develop simplified dynamic and mechanistic models Model dynamic motions of chaperonin Simplify by treating domains as rigid bodies ATP hydrolysis, substrate binding and release Develop meso-scale model of complex Retain mechanical properties Simpler than all-atom representation

Acknowledgements • • • Michael Levitt Sergio Moreno Chris Summa Michael Sykes Dahlia Weiss • Patrice Koehl (UC Davis) • Erik Lindahl (Stockholm University) • Rachel Kolodny (Columbia) Thank You
- Slides: 18