Designing Useful Viruses Steven Skiena Dept of Computer























- Slides: 70

Designing Useful Viruses Steven Skiena Dept. of Computer Science Stony Brook University http: //www. cs. sunysb. edu/~skiena

How might we rapidly create vaccines for new pathogens?

Synthetic Attenuated Virus Engineering (SAVE) Motivation: viral diseases like SARS, 1918 influenza; bioterrorism Input: the genome sequence of a virus Output: a synthetic, attenuated, variant of the virus designed to generate immune response and serve as a vaccine.

Outline of Talk DNA Translation and the Triplet Code Exploiting Redundancy in the Genetic Code Vaccines and Poliovirus Experiments with SAVE Future Work

DNA to RNA to Protein DNA sequences act as templates for building proteins according to the triplet code

The Triplet Code

Which Encoding is Best? There are roughly 3^n possible gene sequences coding for any n-amino acid protein, e. g. 10^75 encodings of a 147 -residue hemoglobin protein. Why did nature select one of them? Alternately, can we exploit this redundancy to design the ‘best’ coding sequence?

What Drives the Evolution of Coding Sequences? Sequences exhibit organism-specific codon bias. Coding helps regulate gene expression with common/scarce codons. RNA secondary structure affects stability. Many signals can be embedded in the coding regions of genes.

Design Criteria for Artificial Genes Matching a given codon/pair distribution Optimizing secondary structure Eliminating or inserting specific patterns Encoding additional gene sequences in alternate reading frames

Incorporating/Excluding Sequence Patterns Many biological features are encoded as substring patterns: restriction sites, mi. RNA targets, stop codons, etc. Differing objectives mandate either including or excluding specific patterns. For example, the restriction enzyme Eco. RI cuts DNA at the pattern GAATTC.

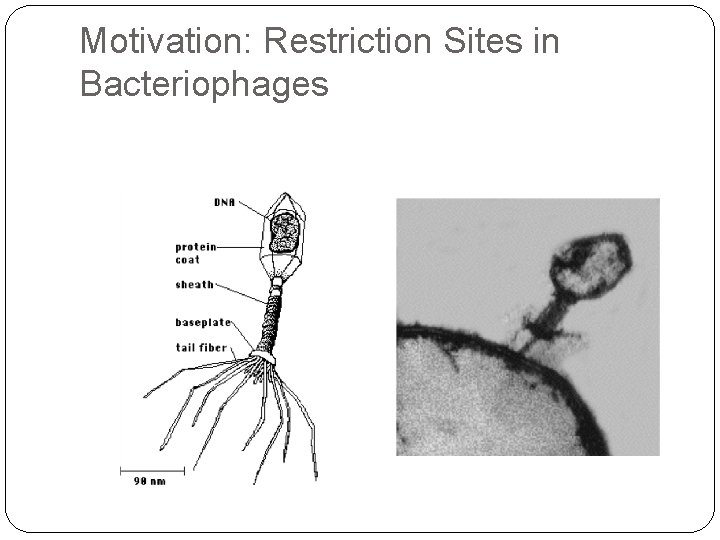
Motivation: Restriction Sites in Bacteriophages

Why Eliminate Restriction Sites? Restriction enzymes exist in bacteria as a defense against phages. Phages have been proposed as an agent against bacterial infections. A theraputic phage might be enhanced by removing all restriction sites from its genome.

Sequence Optimization Algorithms (S. ‘ 01) Dynamic programming can be used to include/exclude many short patterns efficiently, in O(n p 4^k). Since the longest known cutter is only 16 bases, this is a tractible computation. It is NP-complete for long patterns with wildcards, but heuristics work. Our algorithms can remove 90% of restriction sites of all known enzymes

Results by Cutter Length

Optimizing Secondary Structure Nucleotides bind to complementary bases (A-C, G-T/U) so as to minimize their energy. Secondary structures affect molecular interactions and stability Our algorithms design genes with prescribed secondary structure while coding for a given protein

The Zucker-Turner RNA Model Dynamic programming optimizes binding energy over different substructures.

Designing Secondary Structure (Cohen and S. ‘ 02) We can adapt the Zucker-Turner recurrence relations to design a coding sequence maximizing secondary structure in O(n^3). Minimizing secondary structure in the model is NP-complete, but heuristics exist Condon’s group employed our algorithms to design DNA code words (DNA 8)

Maximizing Secondary Structure

How Much Freedom does Nature have for Secondary Structure?

Encoding Genes in Alternate Reading Frames In theory, six coding sequences/ORFs can co- exist on a single DNA sequence. In reality, many viruses do encode overlapping genes to: Reduce genome size Facilitate co-expression

Long Overlaps Exist in Viruses

Compression Algorithm (WPMS ’ 06) Worst case quadratic time Expected time linear because overlaps are usually short

Two arbitrary proteins cannot be significantly interleaved… Overlapping genes in viruses evolved by losing stop codons, not design

… Unless we are free to replace amino acids with similar residues

Why might we want to design overlapping genes? Inserting new genes in a bacterial host is fundamental to biotechnology But the host doesn’t need these genes and deletes them. Interleaving an antibiotic resistance gene in means we can select hosts with the target. There seems to be enough flexibility to make this work.

Chemical Synthesis of Poliovirus Cello et al. synthesized poliovirus c. DNA de novo without a natural template This groundbreaking study made international headlines in July 2002 and opened the new field of synthetic virology. Molla et al. Science. 1991; 254(5038): 1647 -51 Cello, et al. Science. 2002 Aug 9; 297(5583): 1016 -8.

Reverse genetics of poliovirus. Cello, Paul & Wimmer, 2002 Molla, Paul & Wimmer, 1991

Synthetic Biology New synthesis technologies facilitate the engineering of novel biological structures and functions But large-scale synthesis promises to revolutionize how natural organisms are studied as well: “what happens if we change this? ”

DNA Synthesis Technologies Short oligos (50 -100 bases) are readily synthesized Long molecules can be constructed by hybridizing short oligos, but takes work We used Blue Heron for synthesis at $1. 60 / base for ~3000 base sequences The cost for synthesis is dropping rapidly, and is now in the range of $0. 60 /base.

Genomes Species (nt) Poliovirus Phage X 174 Page T 7 “refactoring” Cello, Paul Wimmer 2002 5, 386 11, 515 of 39, 937 Smith et al. 2003 Chan, Kosuti, Endy 2005 “Phoenix” (fossil) progenitor of hum. endog. retrov HERV-K (same as Phoenix) SIVcpz Mycoplasma genitalium reference 7, 500 1918 Influenza virus Human coronavirus (SARS) length 13, 500 9, 472 9, 912 29, 700 582, 970 Tumpey et al. 2005 Dewannieux et al. , 2006 Lee & Bienniaz 2007 Takehisa et al. 2007 Donaldson et al. , 2008 Gibson et al. 2008

Genome-Scale Synthesis? Human genome Chlamydia Mycoplasma pneumoniae Mycoplasma genitalium M. gen. minimal genome 3, 000, 000 1, 226, 265 816, 000 580, 074 ~300, 000 Smallpox virus 185, 570 SARS corona virus 29, 750 Ebola virus 19, 000 1918 Influenza virus*** 13, 500 Yellow fever virus 10, 800 Poliovirus* 7, 500 Phage X 174 (virus of bacteria)** 5, 386 Hepatitis B virus 3, 180 *2002; **2003; ***2005

The Gang: Dimitris Papamichail, Steffan Mueller, Eckard Wimmer, S. , Bruce Futcher, Rob Coleman

RNA viruses Poliovirus is in the Picornaviridae family, (+) stranded, non-enveloped, RNA viruses are the largest virus group, containing dreaded human pathogens (HIV, Ebola, SARS, Dengue, Hanta, Influenza) High mutation rate (1/10, 000 bases) confers high adapability to changing conditions

C 332, 652 H 492, 388 N 98, 245 O 131, 196 P 7, 501 S 2, 340

Poliovirus Genome and Polyprotein Processing 5’ NTR Structural Region Non-structural Region P 1 Cloverleaf VP 4 VP 2 VP 3 P 2 VP 1 IRES P 1 Primary processing Mature proteins VP 4 2 A VP 2 VP 3 VP 1 Structural capsid proteins P 3 2 B 2 A 2 A 3’ NTR 3 A 2 C 3 B P 2 2 B 3 C 7. 5 kb 3 D A A An P 3 2 C 3 A 3 B 3 C 3 D Nonstructural proteins Utilizes IRES in 5’NTR to initiate translation of a single open reading frame Viral proteins produced by cis catalyzed cleavage events Poliovirus genome only 7. 5 kb in length. adapt. Wang, C. .

Jonas Salk (1914 - 1995) Inactivated vaccine (by injection) XXXXXXX xxxxxxxx Albert Sabin (1906 - 1993) attenuated, live vaccine (orally)

Polio Eradication Progress 1988 - 2003 From >125 countries to 6 2003: 784 cases, 6 countries

New Polio Vaccines? Eradication of polio is likely impossible with the current live vaccine because of reversion. WHO has called for a new polio vaccine. Still, our experiments with poliovirus are intended as a proof-of-concept with a well-understood system.

Difficulties in Vaccine Design Few attenuating mutations each having a large effect can easily revert to virulence Function of attenuating mutations poorly defined or not understood at all Attenuation via passaging is costly and time consuming… The poliovirus vaccine strain Sabin 1 was derived by 52 rounds of monkey infections and 16 rounds of monkey kidney cell culture passages, requiring several years of work at prohibitive cost (A total of over 100, 000 monkeys @ $10, 000 = $ 1 Billion)

Synthetic Attenuated Virus Engineering (SAVE) We seek to design a virus which cannot revert by adding large number of mutations each of which is weakly detrimental We seek to deoptimize the genome by interfering with translation while expressing exactly the same proteins (to generate antibody response)

Species-Specific Codon Bias Synonymous codons are used at unequal frequencies Rarely used codons = rare t. RNAs = inhibition of protein translation Replacing unfavorable (rare) codons with favorable synonymous codons leads to improved translation There is some evidence of tissue specific codon bias

Codon Bias Designs n n n Our polio capsid design (PV-AB) n Encoded the same amino-acid sequence n Used only the least frequent codon for each amino-acid in human brain specific genes (and in human tissues in general). Total number of silent mutations: 680 Our polio capsid design (PV-SD) maximized the Hamming distance of the capsid encoding, while keeping the same codon frequency distribution. n Total number of silent mutations: 934 We altered only the capsid coding region because it contains no cisacting structural RNA elements

Codon Alteration Sequence Design To achieve maximum Hamming distance without altering codon bias, we used maximum weight bipartite matching between codon positions and codons, using as weight the number of bases changed. Restriction sites were inserted uniquely (inserted in specific areas and then eliminated everywhere else). Certain regions were locked to preserve secondary structure. Evaluation of secondary structure:

Codon use statistics in PV(M), PV-SD, and PV-AB PV(M) PV-SD PV-AB

Translation of Codon-Bias Designs The “shuffled” polio design translates relatively well despite 534 synonymous changes and is as potent in killing mice as the wildtype. The brain-hostile design translates minimally, but use of smaller segments leads to attenuated strains.

Codon de-optimized Viruses are marked by dramatically reduced infectious virus titers Growth Kinetics on He. La cells titer PFU/ml 1010 PV-wt PV-SD PV-AB 2954 -3386 PV-AB 755 -1513 PV-AB 2470 -2954 * 109 108 107 106 105 104 0 5 10 hrs p. i 15 20 25 *expressed as FFU (focus forming units)

Despite a low titer (biological activity) similar physical amounts of virus particles are produced by codon de-optimized viruses virus PV(M) PV-AB 755 -1513 PV-AB 2470 -2954 virus particles PFU(*FFU) (OD 260 nm) 3. 4 x 1010 9. 4 x 108 1. 04 x 107 4. 24 x 1012 3. 17 x 1012 1. 54 x 1012 virus particles (ELISA) 3. 6 x 1012 2. 1 x 1012 6. 5 x 1011 PFU(*FFU)/ particle ratio 1/115 1/2803 1/105288

equal number of virus particles (virions) Virus A Virus B plaque assay (measures infectious virus titer) many plaques high PFU/particle ratio = high specific infectivity few plaques low PFU/particle ratio = low specific infectivity

Codon-Pair Bias Certain pairs of synonymous codons for two given amino acids are found adjacent to one another more (less) frequently than should be expected. Statistically significant codon-pair bias has been observed in all annotated human genes and other organisms The mechanisms behind this are still unclear, but we can use it to design attenuated viruses. We measure bias with

Codon-Pair Bias is conserved across species

Codon-Pair Bias Designs Codon-pair optimization is essentially the traveling salesman problem We use simulated annealing to shuffle the wildtype codons We produced two designs, maximizing (Max. P 1) and minimizing (Min. P 1) codon pair scores, respectively 1 M P G G P G 18 Original Sequence CPB “Altered” Sequence

Human Genome Codon-Pair Bias Codon pair bias PV-Max: over represented codon pairs (566 silent mutations) PV-Min: under represented codon pairs (631 silent mutations) Codon usage and amino acid sequences of all viruses constant Adapt. Fig. by D. Papamichail

Codon-Pair Bias Sequence Design procedure: Same codon frequency distribution Optimized codon pair score Restriction site uniqueness and elimination Secondary structure folding energy minimization Splice site elimination Goals achieved with simulated annealing, optimization passes and manual intervention.


Growth Kinetics of Synthetic Viruses Display similar kinetics yielding a similar quantity of particles with decreased infectivity (PFU = Plaque Forming Units)

Reduced Specific Infectivity of Codon Pair Bias altered viruses a A 260 - determines particles/ml ® 9. 4 x 1012 particles/ml = 1 A 260 unit b Calculated by dividing the PFU/ml of purified virus by the Particles/ml PFU = Plaque Forming Units

Attenuation of codon pair de-optimized poliovirus correlates with poor translation 80 PV-Min. XY 60 PV-Min 40 PV wt 20 0 viability +++ + + +++ PV-Min. Z PV-Max HCV IRES R-Luc P 1 F-Luc P 2 P 3 AAAn PV IRES F-Luc correlates with the translatabilty of the fused P 1 120 1513 relative F-luc activity % 100 755 2470

Vaccine Experiments

Our designs serve as effective vaccines against PVM-wt Virus Survive Challenge 106 PFU PVM-wt AB 2470 -2954 7/7 Min. XY 7/7 Min. Z 7/7 unvaccinated 1/7

Codon pair de-optimized polioviruses are neuro-attenuated in CD 155 tg mice Virus PLD 50(virions)* PV(M)-wt PV-Max PV-Min. Y PV-Min. XY PV-Min. Z 104. 0 104. 1 105. 0 107. 1 107. 3 PV-Max is NOT a monster virus! * i. c. infections

Synthetic viruses induced neutralizing antibody and protected from lethal challenge Vaccine Protected PV-Min. Z 7/7 PV-Min. XY 7/7 Mock 0/7

Which is responsible for virus attenuation? Cp. G content CPB PV-WT PV-CGhi PV-CPlo 97 216 97 -0. 034 -0. 037 -0. 31 Molly Arabov, unpublished results

Molly Arabov, unpublished results Growth Curve, MOI =3 Specific Infectivity PV-WT 0. 0075 PV-CGhi 0. 0022 PVCPlo 0. 0003 Unpopular codon pairs are worse than many CG pairs

Current and Future Work Experiments with influenza Other design approaches for attenuation Design tools for synthetic biology Experiments with overlapping gene designs

Future work – Sequence design tools

Compressed Gene Designs: Work in Progress We are currently designing an overlapped gene design for synthesis and evaluation Our goal is the “world’s shortest gene’’: a protein complex of n amino acids coded using less than 3 n nucleotides. (with David Green)

Thanks Dimitris Papamichail, Barry Cohen, Bei Wang Steffen Mueller, Rob Coleman, Bruce Futcher, Eckard Wimmer David Green Support from NSF, NIH, Microsoft

Publications • • • Designing Better Phages, S. Skiena, Bioinformatics 17 (2001) S 253 -261. Also ISMB 2001 Natural selection and algorithmic design of m. RNA B. Cohen and S. Skiena, J. Computational Biology 10 (2003) 419 -432 and RECOMB 2002 Two proteins for the price of one: The design of maximally compressed coding sequences B. Wang, D. Papamichail, S. Mueller and S. Skiena, 11 th International Meeting on DNA Computing (DNA 11) 2005 and Lecture Notes in Computer Science 2006, Vol. 3892, pp. 387 -398 Reduction of the rate of poliovirus protein synthesis through large scale codon deoptimization causes virus attenuation of viral virulence S. Mueller, D. Papamichail, J. R. Coleman, S. Skiena and E. Wimmer, Journal of Virology, October 2006, p. 9687 -9696, Vol. 80, No. 19 Synthetic Biology: Synthesis and Modification of a Chemical Called Poliovirus S. Mueller, D. Papamichail, J. Coleman, J. Cello, A. Paul, S. Skiena and E. Wimmer), Future Trends in Microelectronics: The Nano, the Giga, the Ultra, and the Bio, Wiley Interscience, 2007. Virus attenuation by genome-scale changes in codon-pair bias J. Coleman, D. Papamichail, , S. Skiena B. Futcher, S. Mueller, and E. Wimmer), Science, July 2008, p. 1784 -1787, Vol. 320, 2008.


Group Meeting June 2006
 Unlike lytic viruses, lysogenic viruses do not
Unlike lytic viruses, lysogenic viruses do not Unlike lytic viruses, lysogenic viruses do not
Unlike lytic viruses, lysogenic viruses do not Computer viruses presentation
Computer viruses presentation Computer design embroidery
Computer design embroidery Why are viruses considered nonliving?
Why are viruses considered nonliving? Bacteriophage characteristics
Bacteriophage characteristics General characteristics of viruses
General characteristics of viruses Viruses
Viruses Lysogenic viruses do not
Lysogenic viruses do not Section 19-3 diseases caused by bacteria and viruses
Section 19-3 diseases caused by bacteria and viruses Cultivation of viruses
Cultivation of viruses Egg inoculation technique
Egg inoculation technique Egrette - chapter 21
Egrette - chapter 21 Chapter 20 viruses and prokaryotes
Chapter 20 viruses and prokaryotes Blood borne viruses
Blood borne viruses Chapter 7 lesson 1 what are bacteria answer key
Chapter 7 lesson 1 what are bacteria answer key Are viruses alive yes or no
Are viruses alive yes or no Importance of viruses
Importance of viruses Lytic infection
Lytic infection General characters of viruses
General characters of viruses Biosynthesis of rna viruses
Biosynthesis of rna viruses Mackay memorial hospital
Mackay memorial hospital Hepatotropic viruses
Hepatotropic viruses Hepatotropic viruses
Hepatotropic viruses Milad haddad
Milad haddad Chapter 20 viruses and prokaryotes
Chapter 20 viruses and prokaryotes Virus taxonomy
Virus taxonomy Chapter 18 section 2 viruses and prions
Chapter 18 section 2 viruses and prions Replication of viruses
Replication of viruses How do viruses differ from living things
How do viruses differ from living things Importance of viruses
Importance of viruses What kingdom do viruses belong to
What kingdom do viruses belong to Best viruses
Best viruses Best viruses
Best viruses Viruses video
Viruses video Properties of viruses
Properties of viruses Are viruses dead or alive
Are viruses dead or alive General properties of viruses
General properties of viruses Virus
Virus Hershey and chase
Hershey and chase Parts of viruses
Parts of viruses Baltimore classification of virus
Baltimore classification of virus General properties of viruses
General properties of viruses General properties of viruses
General properties of viruses Charateristics of viruses
Charateristics of viruses Section 1 studying viruses and prokaryotes
Section 1 studying viruses and prokaryotes How active viruses multiply
How active viruses multiply Smallest infectious agents
Smallest infectious agents Are viruses decomposers
Are viruses decomposers Dept nmr spectroscopy
Dept nmr spectroscopy Florida dept of agriculture and consumer services
Florida dept of agriculture and consumer services Finance department organizational chart
Finance department organizational chart Building department worcester ma
Building department worcester ma Dept. name of organization (of affiliation)
Dept. name of organization (of affiliation) Mn dept of education
Mn dept of education Dept of finance and administration
Dept of finance and administration Dept. name of organization
Dept. name of organization Ohio dept of dd
Ohio dept of dd Hjdkdkd
Hjdkdkd Vaginal dept
Vaginal dept Gome dept
Gome dept Gome dept
Gome dept Gome dept
Gome dept Gome dept
Gome dept Hoe dept
Hoe dept Fire dept interview questions
Fire dept interview questions Maine dept of agriculture
Maine dept of agriculture Dept of education
Dept of education Florida dept of agriculture and consumer services
Florida dept of agriculture and consumer services Florida dept of agriculture and consumer services
Florida dept of agriculture and consumer services Dept a
Dept a