Design of PPI Modulators for Antiinflammation and Antiinfective




- Slides: 27

Design of PPI Modulators for Anti-inflammation and Anti-infective Research May 2017

Overview • • • Protein-protein interactions (PPIs) represent a vast class of therapeutic targets both inside and outside the cell Protein–protein interactions (PPIs) are involved in most cellular processes and influence biological functions - enzymatic activity, subcellular localization, and/or binding properties The human interactome has been estimated to cover ~400, 000 protein–protein interactions High-resolution structures showed PPI interfaces are generally flat and large (roughly 1000– 2000 A 2 per side) and considered difficult to target Mutational analysis of protein interfaces showed that not all residues at the PPI interface were critical but rather small “hot spots” conferred most of the binding energy (Arkin and Wells, 2004; Clackson and Wells, 1995). Hot spots tended to cluster at the center of the interface, to cover an area comparable to the size of a small molecule, to be hydrophobic, and to show conformational adaptivity Modulation of PPIs provide a wealth of opportunities for therapeutic intervention in a broad range of disease conditions

Types of PPI by Interaction Small interface and short peptide sequence for interaction Partner proteins with secondary structure binding to a hydrophobic groove Complex structure of interaction on both sides, multiple points of interaction • • PPI come in many shapes and sizes Most of the clinical-stage inhibitors target PPI primary interaction where the hot-spot residues are concentrated in small binding pockets (250 – 900 A 2) and partner proteins are characterized by short primary sequences at the interface PPI that contain a single unit of secondary structure, such as an alpha helix, binding to a hydrophobic groove Globular interfaces, requiring tertiary structure on both sides of the PPI

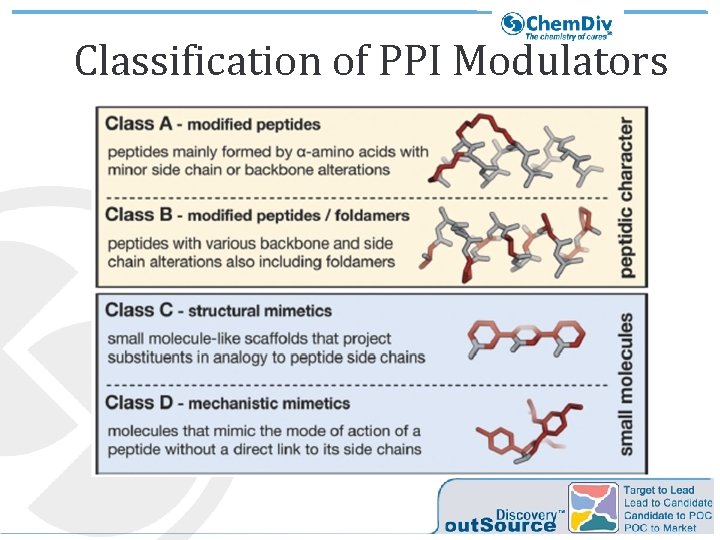
Classification of PPI Modulators

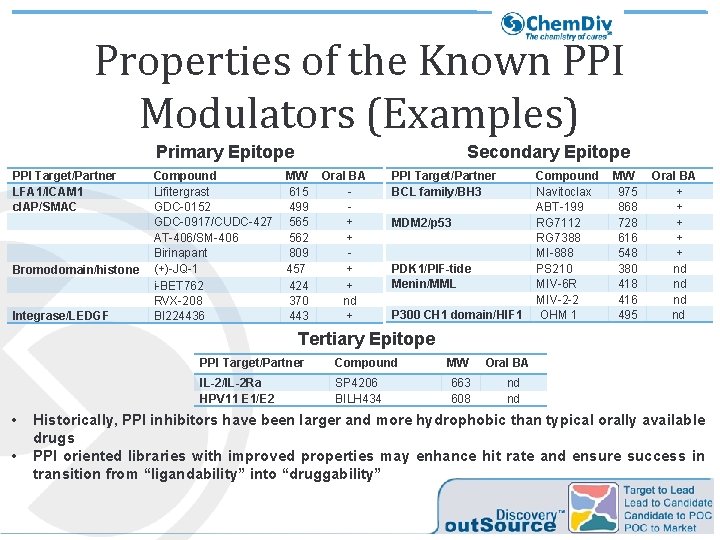
Properties of the Known PPI Modulators (Examples) Primary Epitope PPI Target/Partner LFA 1/ICAM 1 c. IAP/SMAC Bromodomain/histone Integrase/LEDGF Compound Lifitergrast GDC-0152 GDC-0917/CUDC-427 AT-406/SM-406 Birinapant (+)-JQ-1 i-BET 762 RVX-208 BI 224436 Secondary Epitope MW 615 499 565 562 809 457 424 370 443 Oral BA + + nd + PPI Target/Partner BCL family/BH 3 MDM 2/p 53 PDK 1/PIF-tide Menin/MML P 300 CH 1 domain/HIF 1 Compound Navitoclax ABT-199 RG 7112 RG 7388 MI-888 PS 210 MIV-6 R MIV-2 -2 OHM 1 MW 975 868 728 616 548 380 418 416 495 Oral BA + + + nd nd Tertiary Epitope • • PPI Target/Partner Compound IL-2/IL-2 Ra HPV 11 E 1/E 2 SP 4206 BILH 434 MW 663 608 Oral BA nd nd Historically, PPI inhibitors have been larger and more hydrophobic than typical orally available drugs PPI oriented libraries with improved properties may enhance hit rate and ensure success in transition from “ligandability” into “druggability”

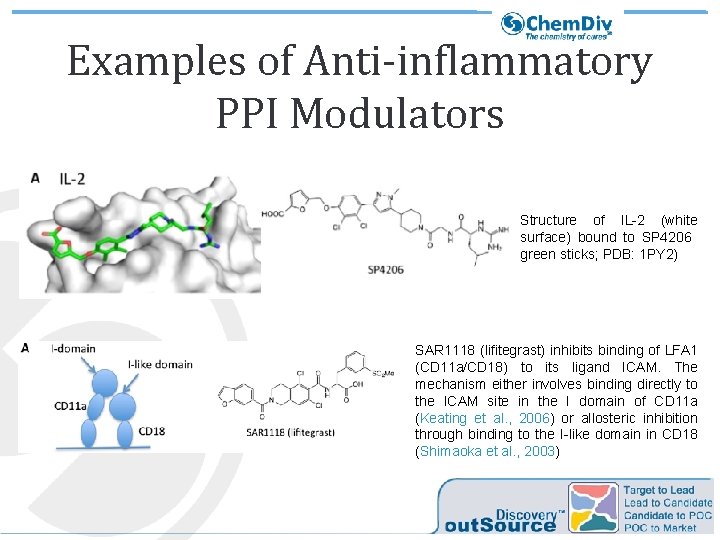
Examples of Anti-inflammatory PPI Modulators Structure of IL-2 (white surface) bound to SP 4206 green sticks; PDB: 1 PY 2) SAR 1118 (lifitegrast) inhibits binding of LFA 1 (CD 11 a/CD 18) to its ligand ICAM. The mechanism either involves binding directly to the ICAM site in the I domain of CD 11 a (Keating et al. , 2006) or allosteric inhibition through binding to the I-like domain in CD 18 (Shimaoka et al. , 2003)

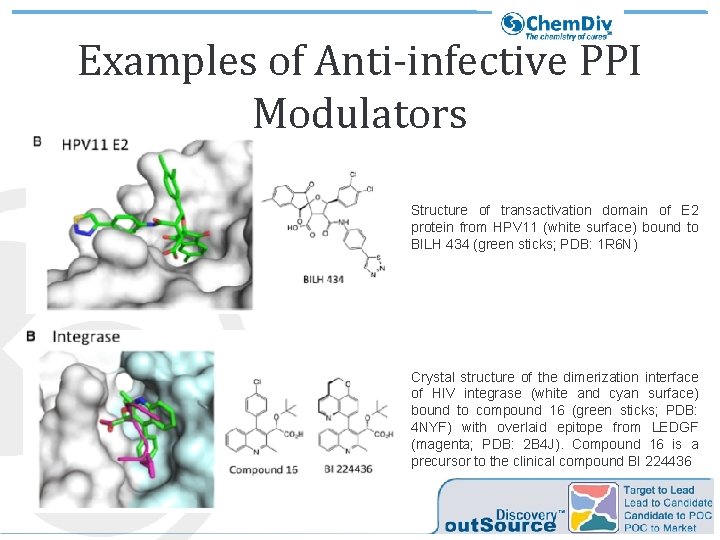
Examples of Anti-infective PPI Modulators Structure of transactivation domain of E 2 protein from HPV 11 (white surface) bound to BILH 434 (green sticks; PDB: 1 R 6 N) Crystal structure of the dimerization interface of HIV integrase (white and cyan surface) bound to compound 16 (green sticks; PDB: 4 NYF) with overlaid epitope from LEDGF (magenta; PDB: 2 B 4 J). Compound 16 is a precursor to the clinical compound BI 224436

Commercial Libraries Targeted at PPIs* Supplier Chem. Div Otava Chemicals Asinex Com. Innex Life Chemicals NQuix No. of Compounds 125, 000** 1, 330 1, 020 520 7, 000 custom 850 23, 200 4, 300 NA Design Method β-turn-, helix-, 3 D-, peptidomimetics, macrocycles, spiro compounds decision trees similarity search β-turn mimetics shape analysis helix mimetics, macrocycles machine learning 2 D fingerprint similarity rule-of-four NA * - Huggins et al; Chem Biol. 2015 Jun 18; 22(6): 689– 703 Overcoming Chemical, Biological, and Computational Challenges in the Development of Inhibitors Targeting Protein-Protein Interactions ** - Library size as published

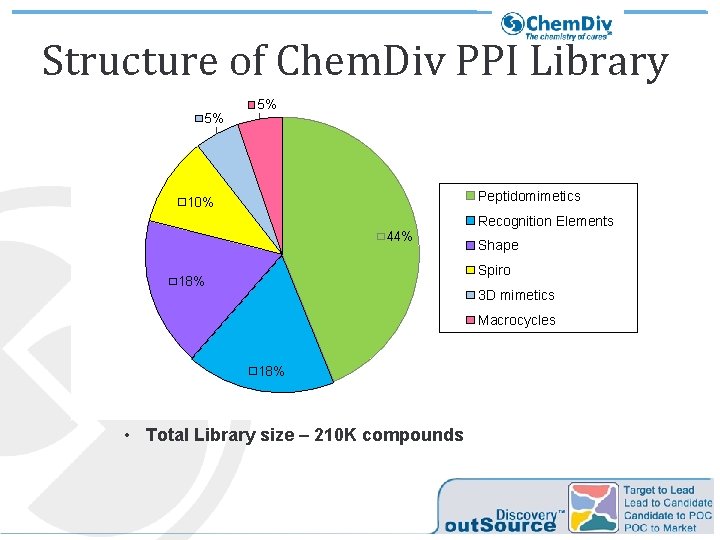
Structure of Chem. Div PPI Library 5% 5% Peptidomimetics 10% Recognition Elements 44% Shape Spiro 18% 3 D mimetics Macrocycles 18% • Total Library size – 210 K compounds

Turn Mimetics and Approaches for Targeting

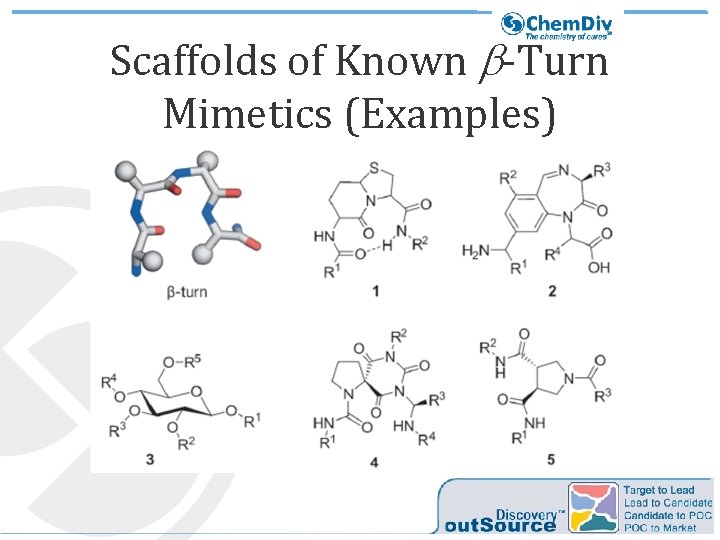
Scaffolds of Known b-Turn Mimetics (Examples)

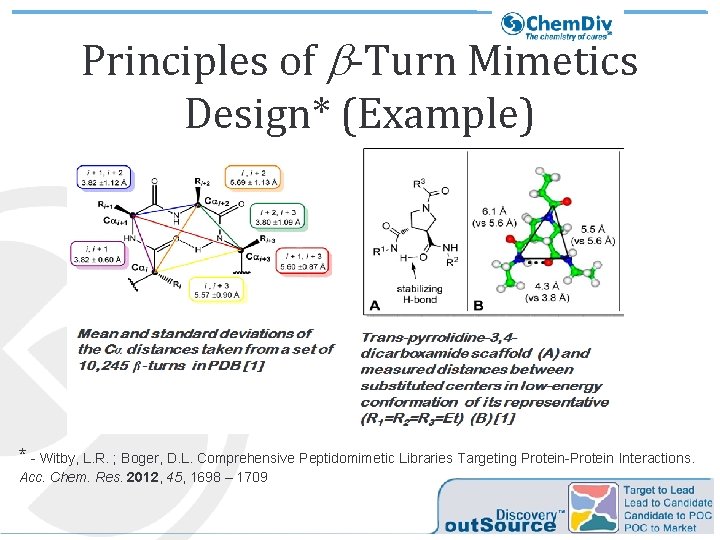
Principles of b-Turn Mimetics Design* (Example) * - Witby, L. R. ; Boger, D. L. Comprehensive Peptidomimetic Libraries Targeting Protein-Protein Interactions. Acc. Chem. Res. 2012, 45, 1698 – 1709

Chem. Div’s b-Turn Mimetics (Examples)

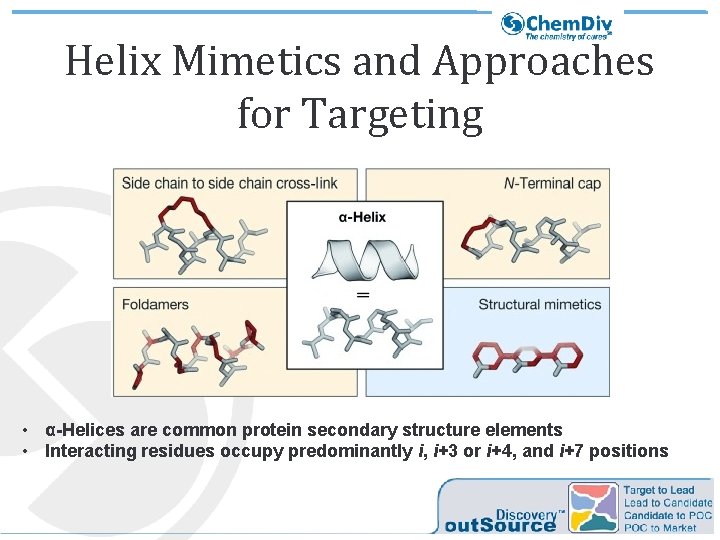
Helix Mimetics and Approaches for Targeting • α-Helices are common protein secondary structure elements • Interacting residues occupy predominantly i, i+3 or i+4, and i+7 positions

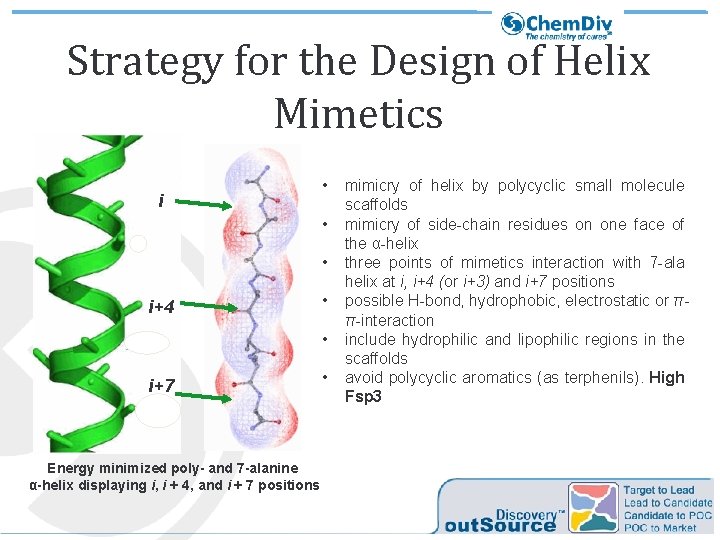
Strategy for the Design of Helix Mimetics i • • • i+4 • • i+7 Energy minimized poly- and 7 -alanine α-helix displaying i, i + 4, and i + 7 positions • mimicry of helix by polycyclic small molecule scaffolds mimicry of side-chain residues on one face of the α-helix three points of mimetics interaction with 7 -ala helix at i, i+4 (or i+3) and i+7 positions possible H-bond, hydrophobic, electrostatic or ππ-interaction include hydrophilic and lipophilic regions in the scaffolds avoid polycyclic aromatics (as terphenils). High Fsp 3

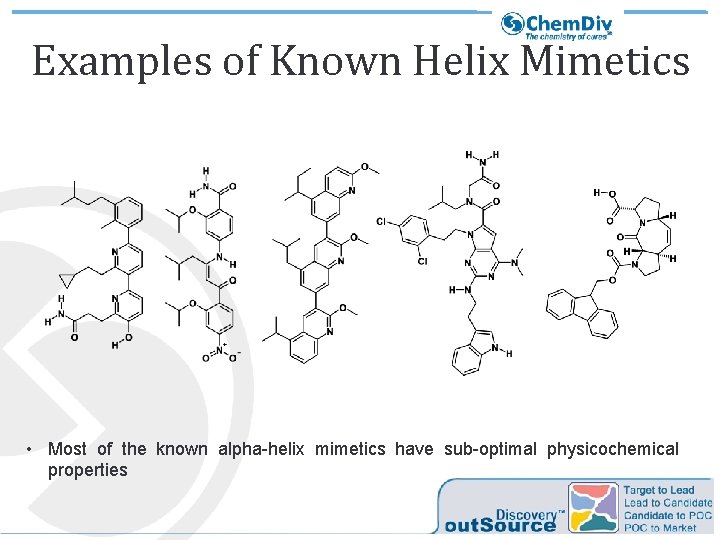
Examples of Known Helix Mimetics • Most of the known alpha-helix mimetics have sub-optimal physicochemical properties

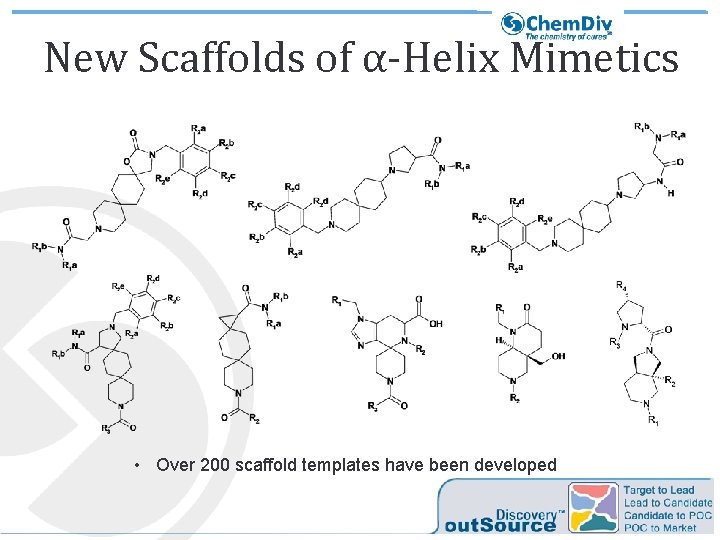
New Scaffolds of α-Helix Mimetics • Over 200 scaffold templates have been developed

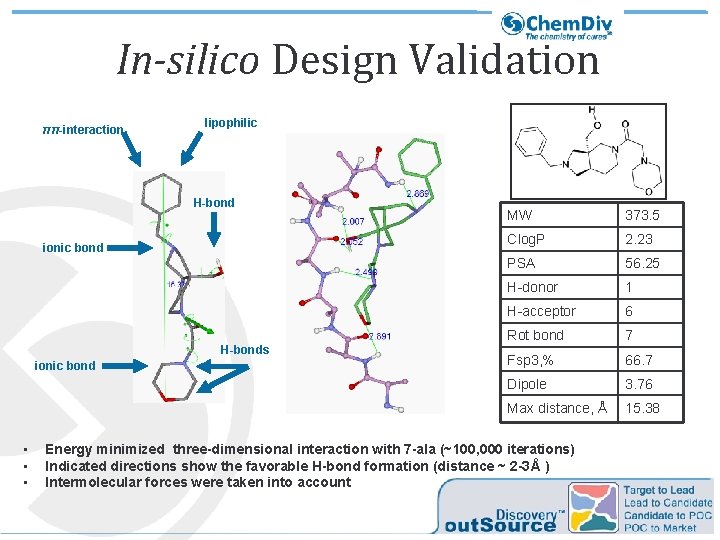
In-silico Design Validation ππ-interaction lipophilic H-bond ionic bond H-bonds ionic bond • • • MW 373. 5 Clog. P 2. 23 PSA 56. 25 H-donor 1 H-acceptor 6 Rot bond 7 Fsp 3, % 66. 7 Dipole 3. 76 Max distance, Å 15. 38 Energy minimized three-dimensional interaction with 7 -ala (~100, 000 iterations) Indicated directions show the favorable H-bond formation (distance ~ 2 -3Å ) Intermolecular forces were taken into account

Macrocyclic as PPI Modulators

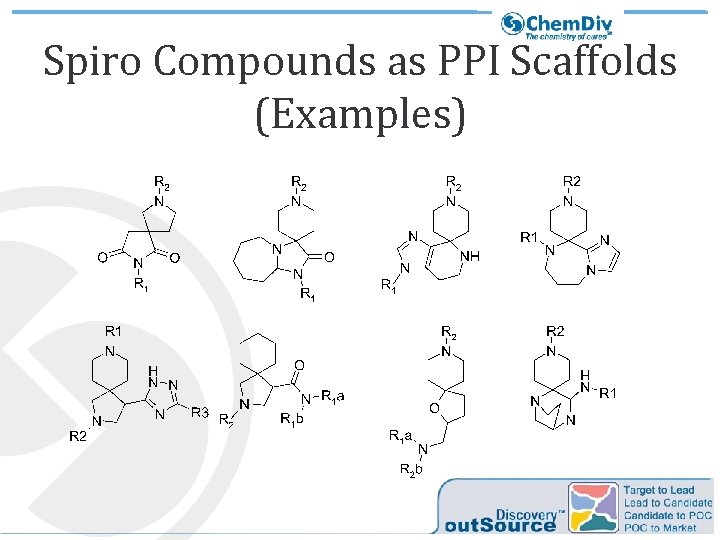
Spiro Compounds as PPI Scaffolds (Examples)

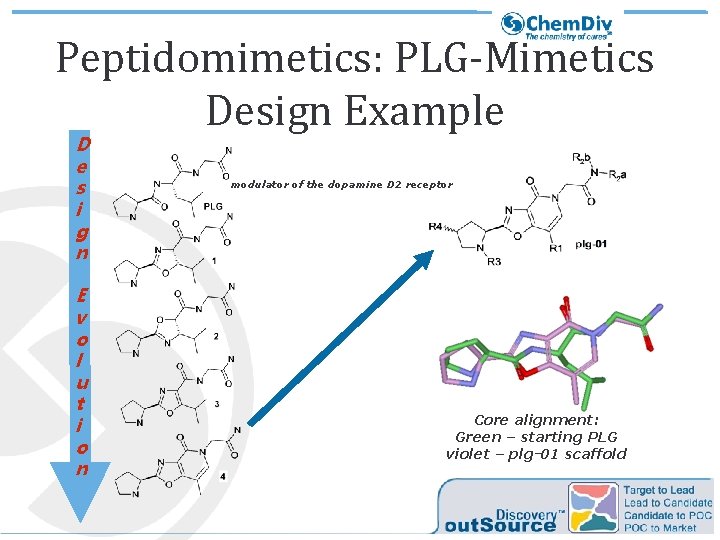
Peptidomimetics: PLG-Mimetics Design Example D e s i g n E v o l u t i o n modulator of the dopamine D 2 receptor Core alignment: Green – starting PLG violet – plg-01 scaffold

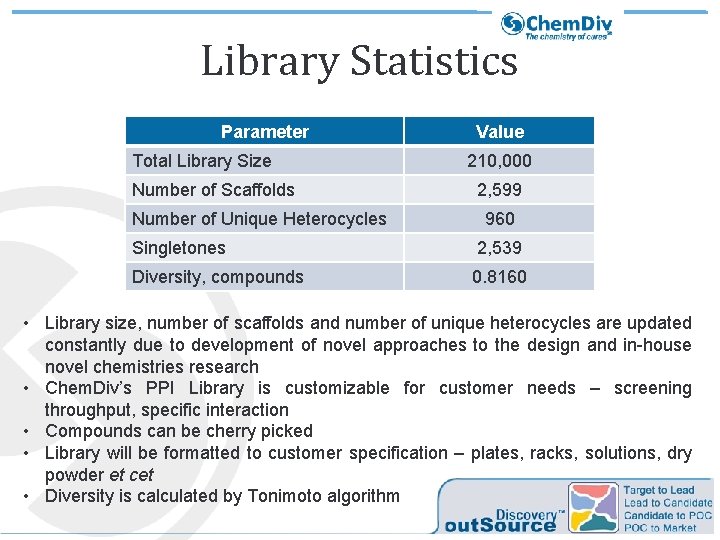
Library Statistics Parameter Total Library Size Number of Scaffolds Number of Unique Heterocycles Value 210, 000 2, 599 960 Singletones 2, 539 Diversity, compounds 0. 8160 • Library size, number of scaffolds and number of unique heterocycles are updated constantly due to development of novel approaches to the design and in-house novel chemistries research • Chem. Div’s PPI Library is customizable for customer needs – screening throughput, specific interaction • Compounds can be cherry picked • Library will be formatted to customer specification – plates, racks, solutions, dry powder et cet • Diversity is calculated by Tonimoto algorithm

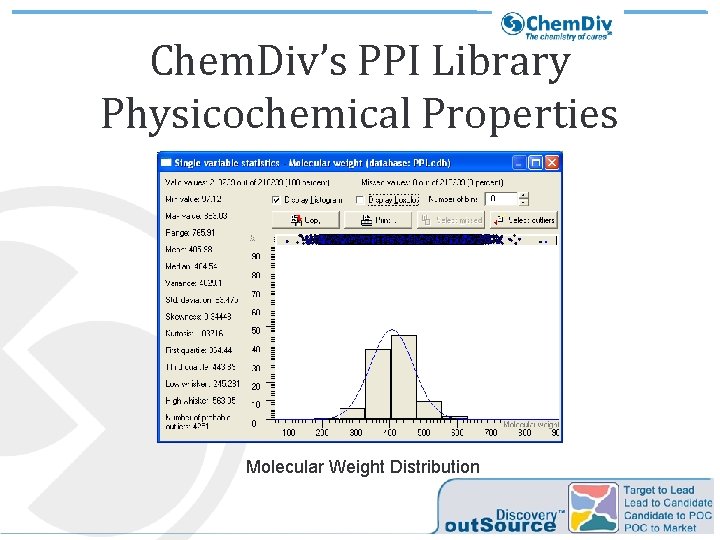
Chem. Div’s PPI Library Physicochemical Properties Molecular Weight Distribution

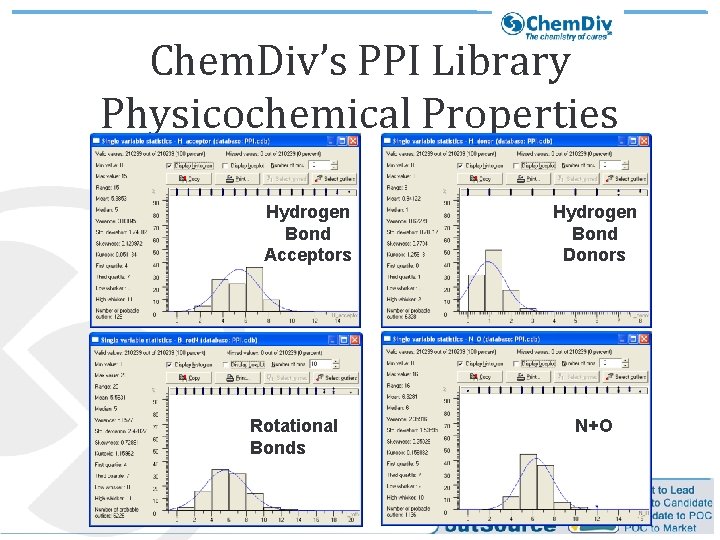
Chem. Div’s PPI Library Physicochemical Properties Hydrogen Bond Acceptors Rotational Bonds Hydrogen Bond Donors N+O

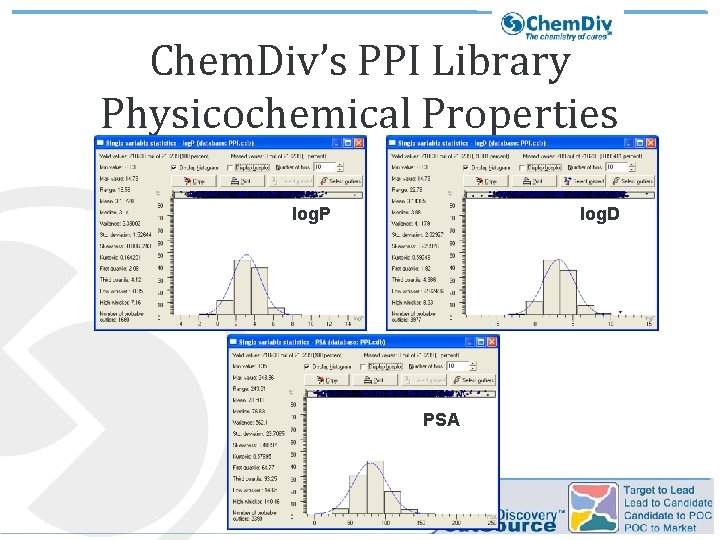
Chem. Div’s PPI Library Physicochemical Properties log. P log. D PSA

Chem. Div’s PPI Library • • Largest commercially available PPI library Designed to cover most epitope, hot-spot interactions Unique chemistry – strong IP potential Customizable to customer needs – – Size Cherry-picking Format options Delivery options • Expandable from stock and synthesis • Attractively priced

Thank you!