DECODING OF EXON SPLICING PATTERNS IN THE HUMAN




- Slides: 14

DECODING OF EXON SPLICING PATTERNS IN THE HUMAN RUNX 1 -RUNX 1 T 1 FUSION GENE Vasily V. Grinev Associate Professor Department of Genetics Faculty of Biology Belarusian State University Minsk, Republic of Belarus

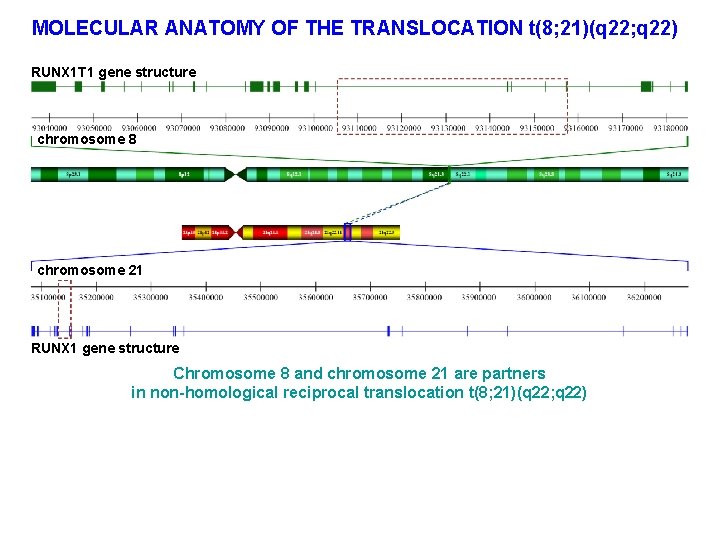
MOLECULAR ANATOMY OF THE TRANSLOCATION t(8; 21)(q 22; q 22) RUNX 1 T 1 gene structure chromosome 8 chromosome 21 RUNX 1 gene structure Chromosome 8 and chromosome 21 are partners in non-homological reciprocal translocation t(8; 21)(q 22; q 22)

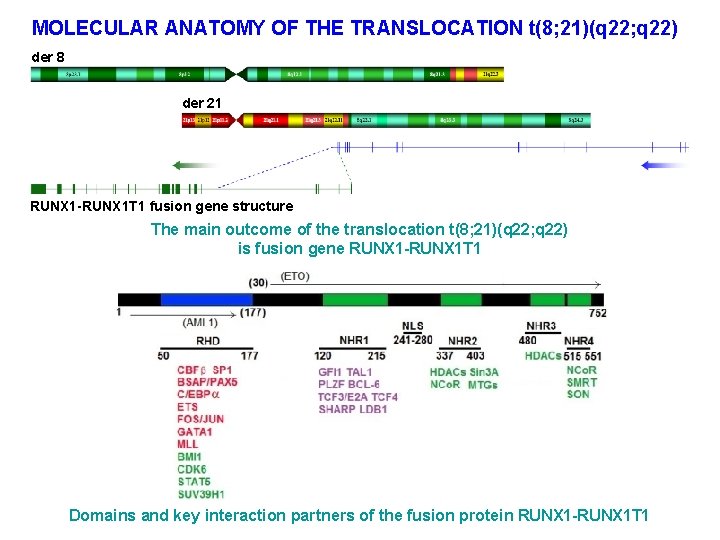
MOLECULAR ANATOMY OF THE TRANSLOCATION t(8; 21)(q 22; q 22) der 8 der 21 RUNX 1 -RUNX 1 T 1 fusion gene structure The main outcome of the translocation t(8; 21)(q 22; q 22) is fusion gene RUNX 1 -RUNX 1 T 1 Domains and key interaction partners of the fusion protein RUNX 1 -RUNX 1 T 1

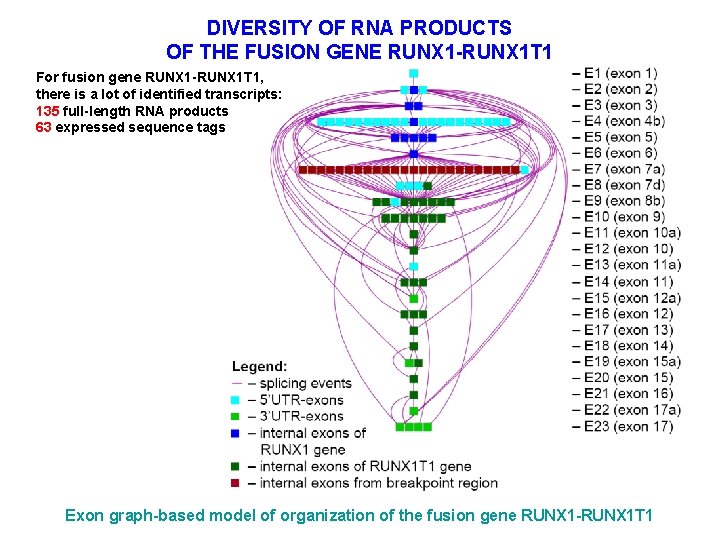
DIVERSITY OF RNA PRODUCTS OF THE FUSION GENE RUNX 1 -RUNX 1 T 1 For fusion gene RUNX 1 -RUNX 1 T 1, there is a lot of identified transcripts: 135 full-length RNA products 63 expressed sequence tags Exon graph-based model of organization of the fusion gene RUNX 1 -RUNX 1 T 1

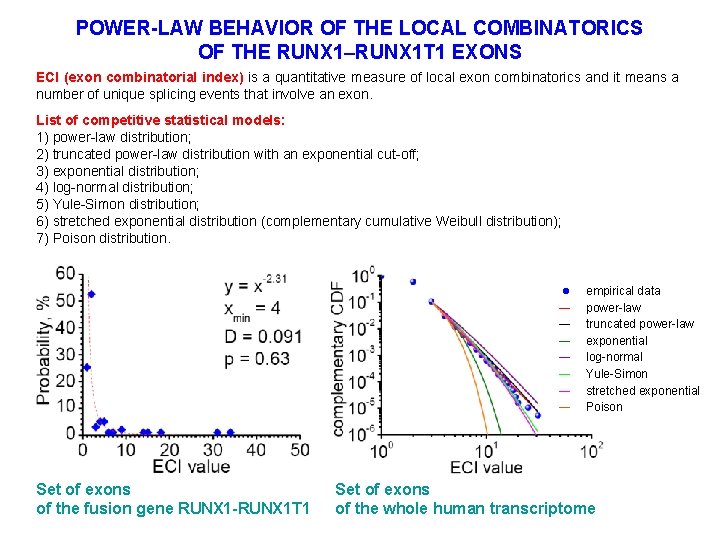
POWER-LAW BEHAVIOR OF THE LOCAL COMBINATORICS OF THE RUNX 1–RUNX 1 T 1 EXONS ECI (exon combinatorial index) is a quantitative measure of local exon combinatorics and it means a number of unique splicing events that involve an exon. List of competitive statistical models: 1) power-law distribution; 2) truncated power-law distribution with an exponential cut-off; 3) exponential distribution; 4) log-normal distribution; 5) Yule-Simon distribution; 6) stretched exponential distribution (complementary cumulative Weibull distribution); 7) Poison distribution. ― ― ― ― Set of exons of the fusion gene RUNX 1 -RUNX 1 T 1 empirical data power-law truncated power-law exponential log-normal Yule-Simon stretched exponential Poison Set of exons of the whole human transcriptome

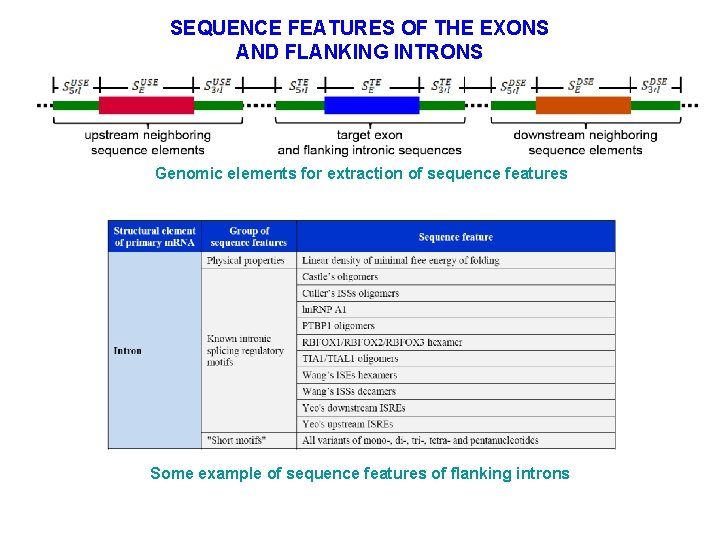
SEQUENCE FEATURES OF THE EXONS AND FLANKING INTRONS Genomic elements for extraction of sequence features Some example of sequence features of flanking introns

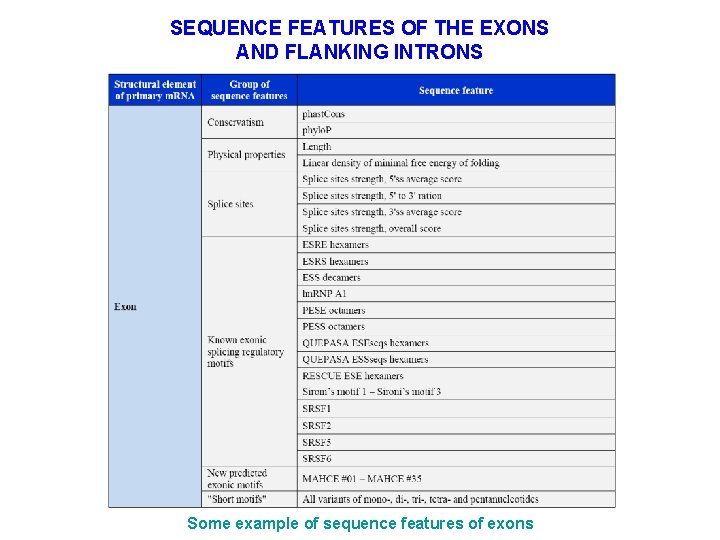
SEQUENCE FEATURES OF THE EXONS AND FLANKING INTRONS Some example of sequence features of exons

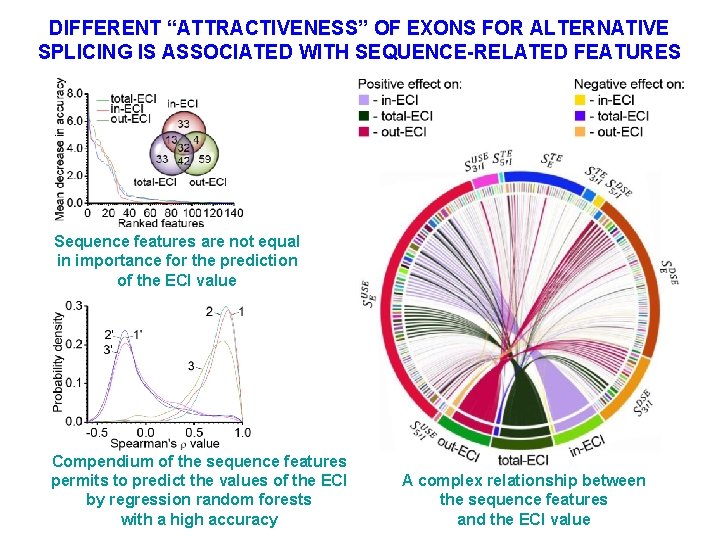
DIFFERENT “ATTRACTIVENESS” OF EXONS FOR ALTERNATIVE SPLICING IS ASSOCIATED WITH SEQUENCE-RELATED FEATURES Sequence features are not equal in importance for the prediction of the ECI value Compendium of the sequence features permits to predict the values of the ECI by regression random forests with a high accuracy A complex relationship between the sequence features and the ECI value

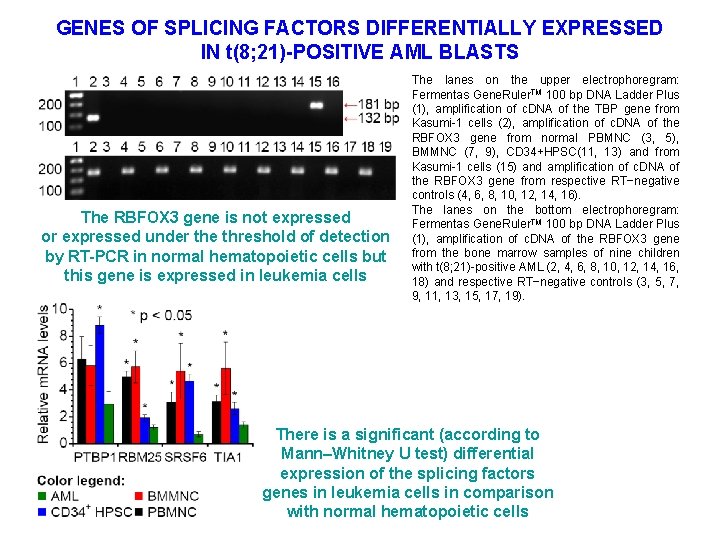
GENES OF SPLICING FACTORS DIFFERENTIALLY EXPRESSED IN t(8; 21)-POSITIVE AML BLASTS The RBFOX 3 gene is not expressed or expressed under the threshold of detection by RT-PCR in normal hematopoietic cells but this gene is expressed in leukemia cells The lanes on the upper electrophoregram: Fermentas Gene. Ruler. TM 100 bp DNA Ladder Plus (1), amplification of c. DNA of the TBP gene from Kasumi-1 cells (2), amplification of c. DNA of the RBFOX 3 gene from normal PBMNC (3, 5), BMMNC (7, 9), CD 34+HPSC(11, 13) and from Kasumi-1 cells (15) and amplification of c. DNA of the RBFOX 3 gene from respective RT−negative controls (4, 6, 8, 10, 12, 14, 16). The lanes on the bottom electrophoregram: Fermentas Gene. Ruler. TM 100 bp DNA Ladder Plus (1), amplification of c. DNA of the RBFOX 3 gene from the bone marrow samples of nine children with t(8; 21)-positive AML (2, 4, 6, 8, 10, 12, 14, 16, 18) and respective RT−negative controls (3, 5, 7, 9, 11, 13, 15, 17, 19). There is a significant (according to Mann–Whitney U test) differential expression of the splicing factors genes in leukemia cells in comparison with normal hematopoietic cells

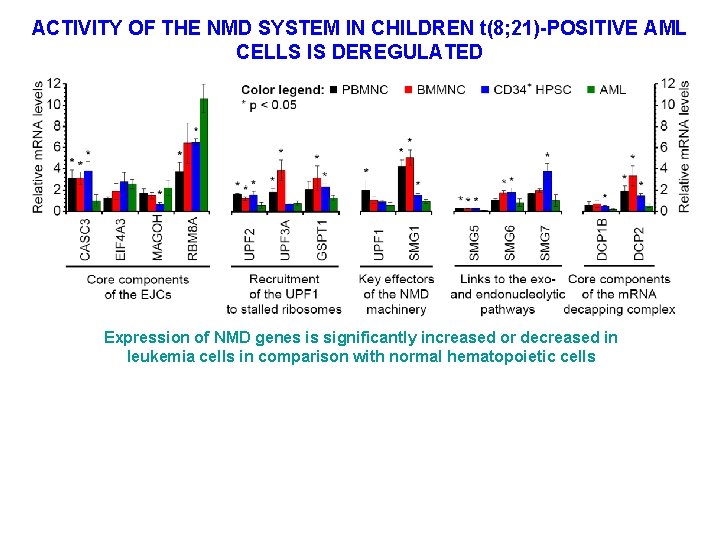
ACTIVITY OF THE NMD SYSTEM IN CHILDREN t(8; 21)-POSITIVE AML CELLS IS DEREGULATED Expression of NMD genes is significantly increased or decreased in leukemia cells in comparison with normal hematopoietic cells

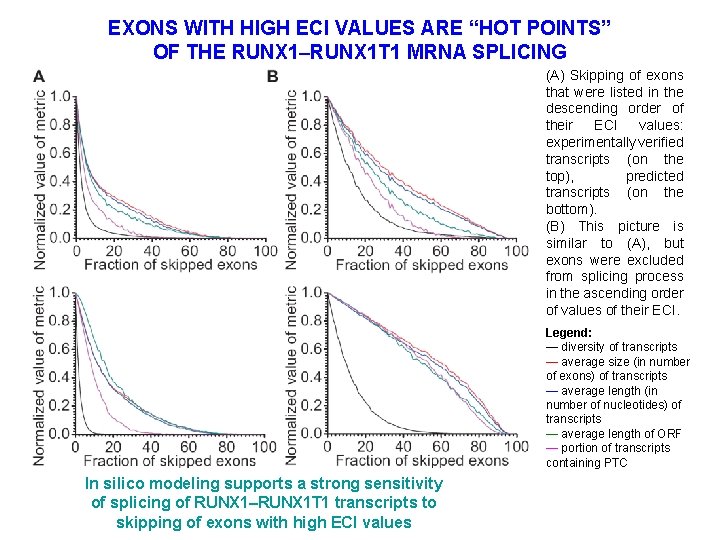
EXONS WITH HIGH ECI VALUES ARE “HOT POINTS” OF THE RUNX 1–RUNX 1 T 1 MRNA SPLICING (A) Skipping of exons that were listed in the descending order of their ECI values: experimentally verified transcripts (on the top), predicted transcripts (on the bottom). (B) This picture is similar to (A), but exons were excluded from splicing process in the ascending order of values of their ECI. Legend: — diversity of transcripts — average size (in number of exons) of transcripts — average length (in number of nucleotides) of transcripts — average length of ORF — portion of transcripts containing PTC In silico modeling supports a strong sensitivity of splicing of RUNX 1–RUNX 1 T 1 transcripts to skipping of exons with high ECI values

MORE DETAILS IN:

MANY THANKS TO THE MEMBERS OF OUR TEAM: Alexandr A. Migas Olga A. Mishkova Olga V. Aleinikova Petr V. Nazarov Laurent Vallar Tatiana V. Ramanouskaya Alina V. Vaitsiankova Ilia M. Ilyushonak Natalia Jr. Siomava Aksana D. Kirsanava

THANK YOU FOR ATTENTION! HETEROZYGOATS Just allele uneven