Deciphering the Prenatal Microarray Alan Ma OG Meeting







- Slides: 29

Deciphering the Prenatal Microarray Alan Ma O&G Meeting 12 th November 2014

Aims • • • Role of microarray in prenatal testing How we decipher the microarray results Common scenarios Unrelated/incidental findings of significance Future directions

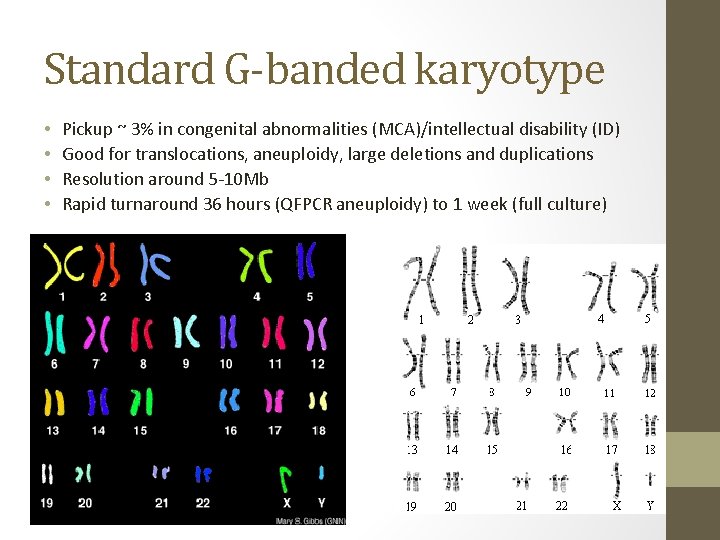
Standard G-banded karyotype • • Pickup ~ 3% in congenital abnormalities (MCA)/intellectual disability (ID) Good for translocations, aneuploidy, large deletions and duplications Resolution around 5 -10 Mb Rapid turnaround 36 hours (QFPCR aneuploidy) to 1 week (full culture)


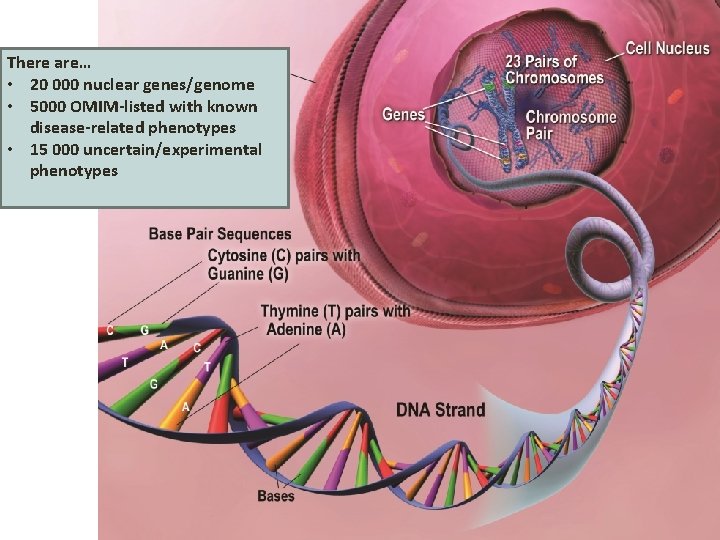
There are… • 20 000 nuclear genes/genome • 5000 OMIM-listed with known disease-related phenotypes • 15 000 uncertain/experimental phenotypes

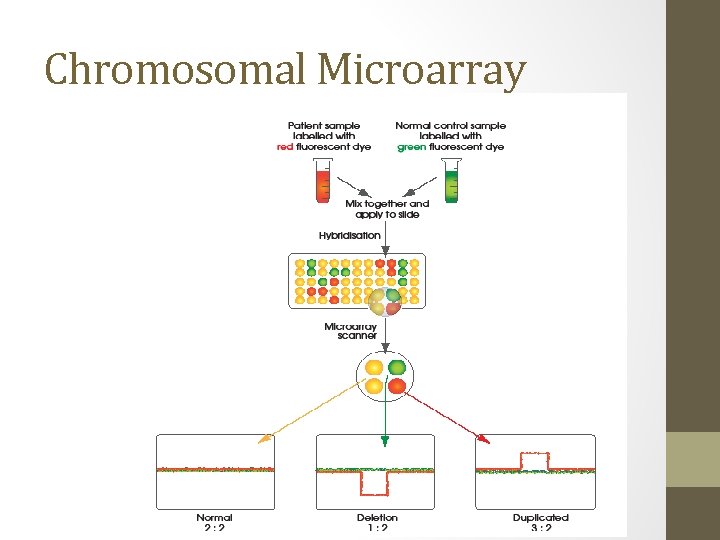
Chromosomal Microarray

Chromosomal Microarray

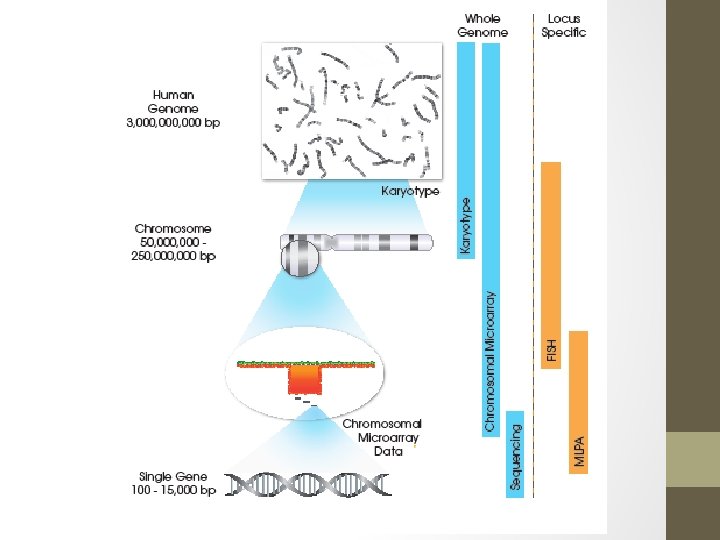
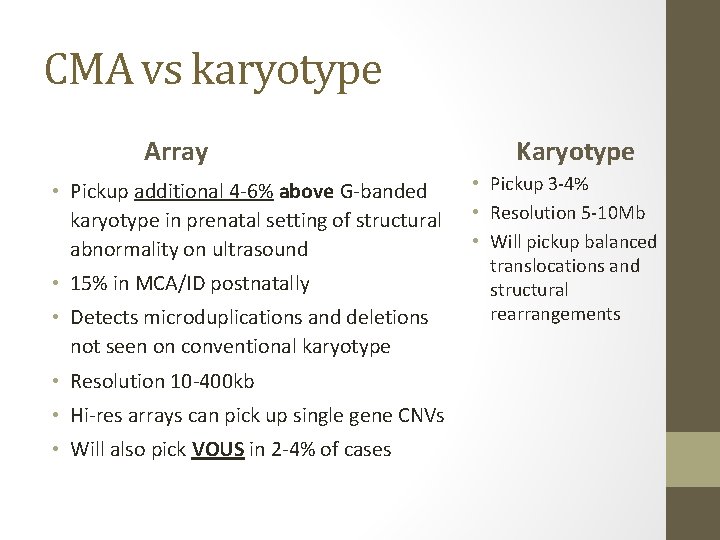
CMA vs karyotype Array • Pickup additional 4 -6% above G-banded karyotype in prenatal setting of structural abnormality on ultrasound • 15% in MCA/ID postnatally • Detects microduplications and deletions not seen on conventional karyotype • Resolution 10 -400 kb • Hi-res arrays can pick up single gene CNVs • Will also pick VOUS in 2 -4% of cases Karyotype • Pickup 3 -4% • Resolution 5 -10 Mb • Will pickup balanced translocations and structural rearrangements

60 K Agilent Array

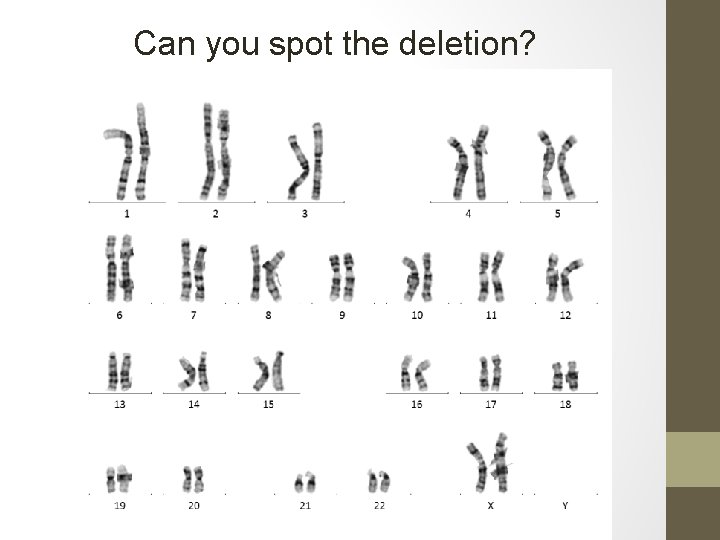
Can you spot the deletion?

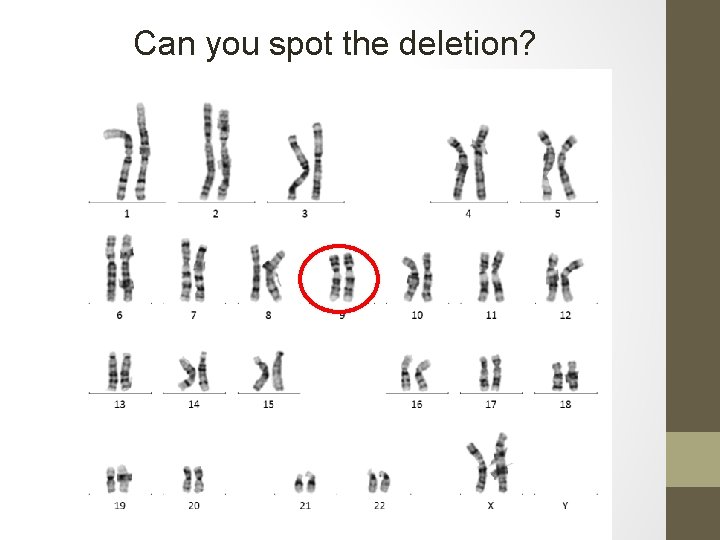
Can you spot the deletion?

Can you spot the deletion?

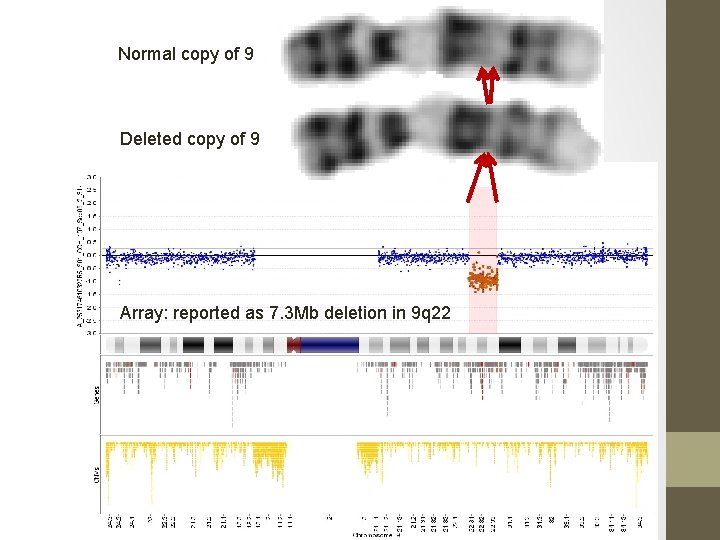
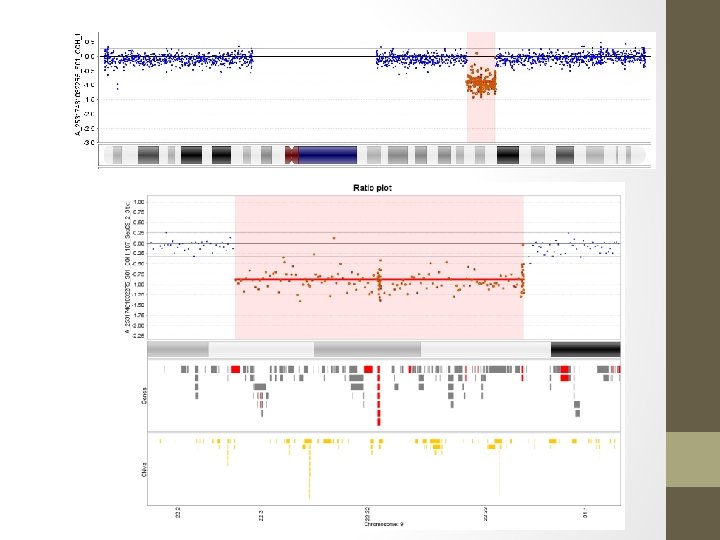
Normal copy of 9 Deleted copy of 9 Array: reported as 7. 3 Mb deletion in 9 q 22



Benefits and disadvantages Benefits • Higher yield • Genome-wide • Delineates deletions/ duplications more clearly – ‘what genes are in there? ’ = more precise answer Disadvantages • Cannot detect balanced translocations or map imbalances • Misses point mutations and small del/dups < 10 kb • Can ‘unmask’ carriers of recessive conditions, unrelated conditions and consanguinuity! • Can be hard to interpret results and counsel - VOUSes • Turnaround time • 2 weeks • Longer if issues with DNA quality/quantity, parental studies

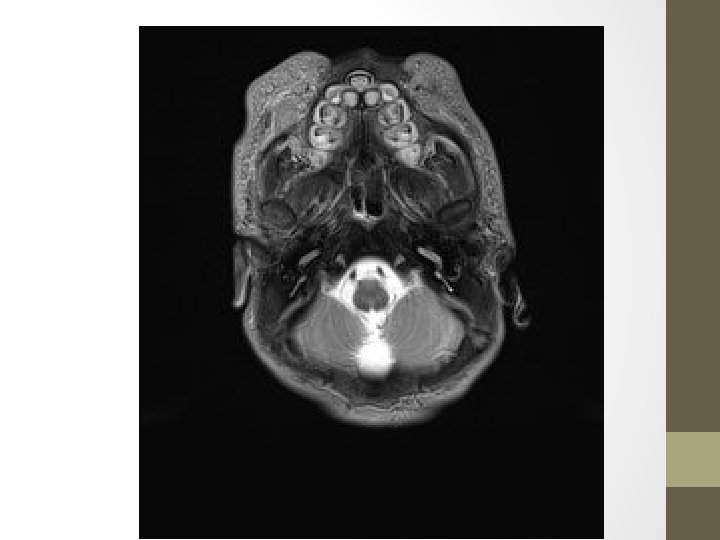
Neonatal consult • Respiratory distress • Nasal piriform aperture stenosis • Hypoteloric and hypotonic


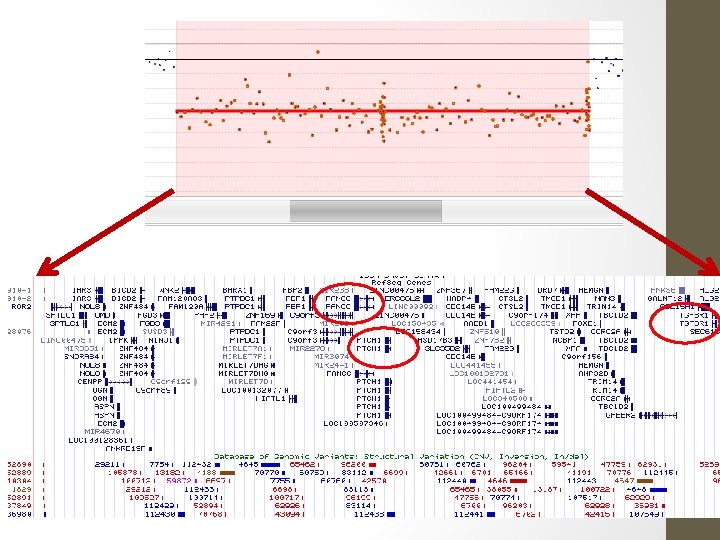
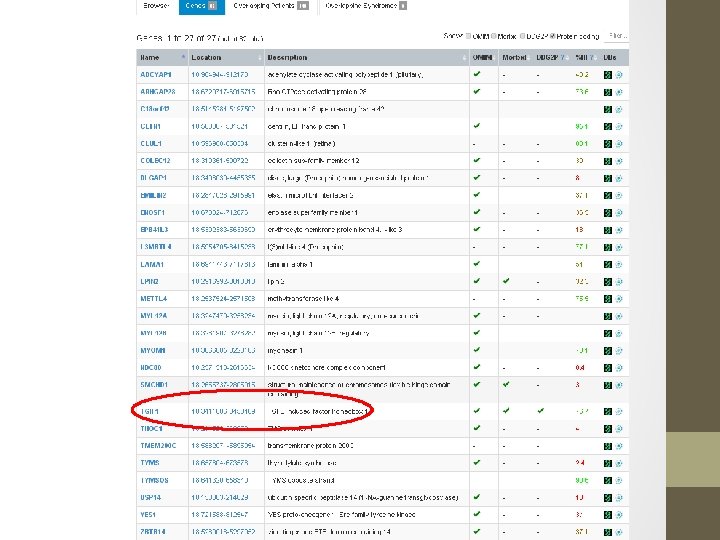
RESULT: A deletion was detected on CGH microarray. The ISCN (2009) description is: arr 18 p 11. 32 p 11. 31(138, 963 -6, 963, 069)x 1 CONCLUSION: Microarray testing detects a terminal deletion, within chromosome 18 bands p 11. 32 to p 11. 31. This has minimum size 6. 82 Mb, and extends from position 0. 14 to 6. 96 Mb. (Max. size is 7. 08 Mb, from 0 to 7. 08). The deletion includes approx. 40 known genes, from ROCK 1 P 1 to LAMA 1. Two of these are associated with OMIM-listed disease, namely LPIN 2 & TGIF 1.

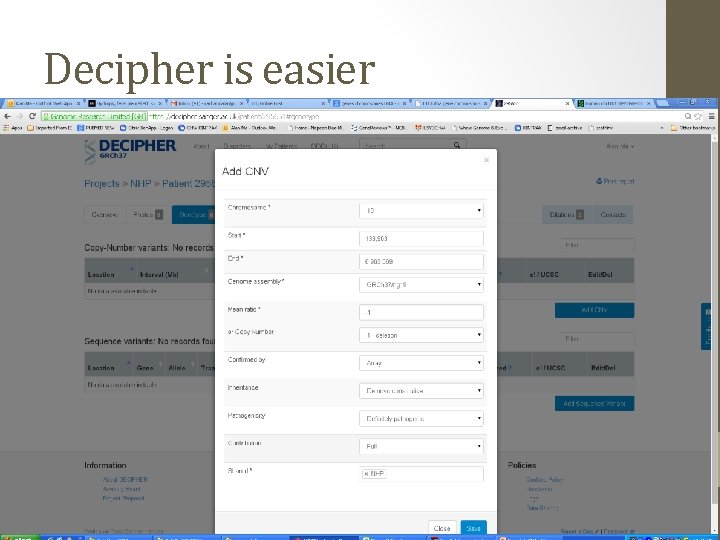
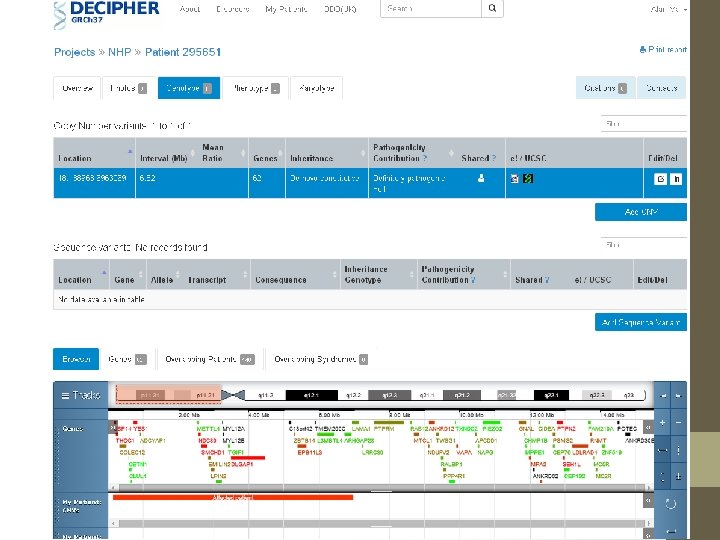
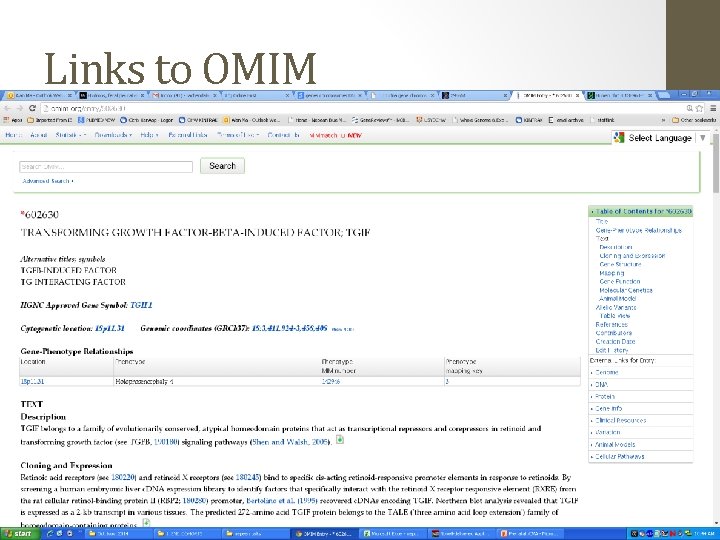
2 ways to analyse this USCS Genome Browser http: //genome. ucsc. edu/ • Free to use • Data-overload Decipher Database https: //decipher. sanger. ac. uk/index • Curated database of CNVs (Wellcome Sanger) • Need membership (easy)

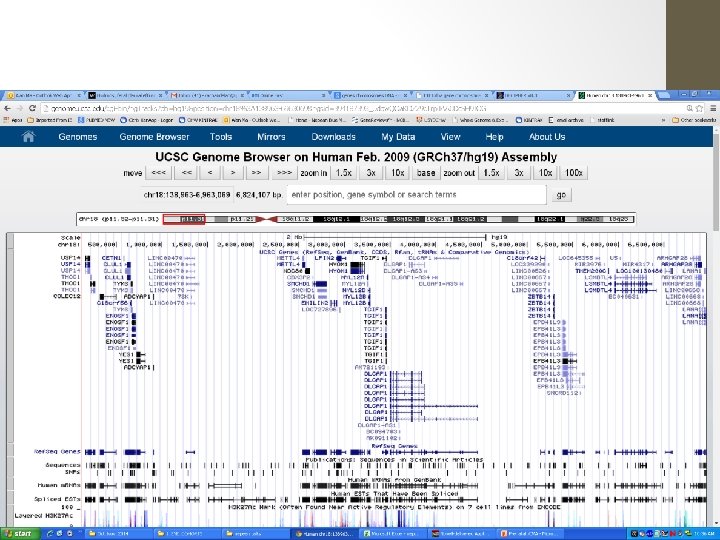
USCS Genome Browser

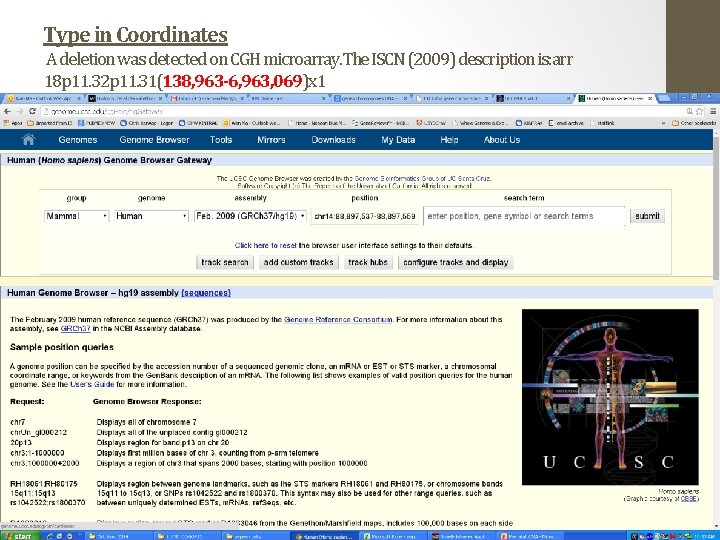
Type in Coordinates A deletion was detected on CGH microarray. The ISCN (2009) description is: arr 18 p 11. 32 p 11. 31(138, 963 -6, 963, 069)x 1

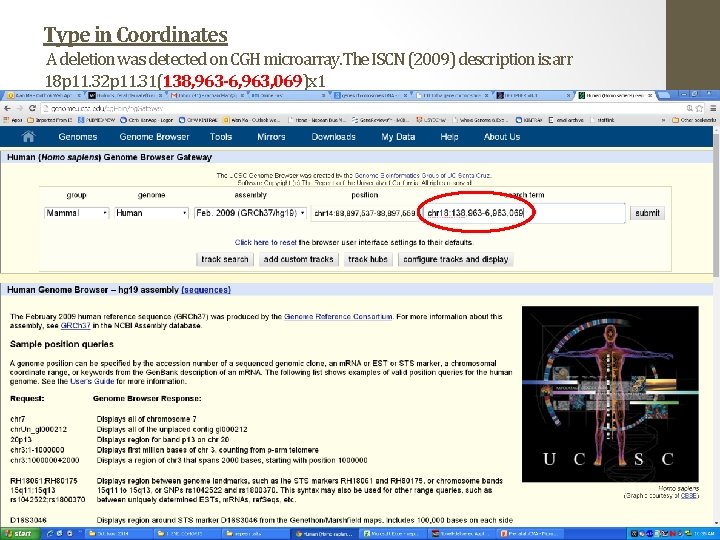
Type in Coordinates A deletion was detected on CGH microarray. The ISCN (2009) description is: arr 18 p 11. 32 p 11. 31(138, 963 -6, 963, 069)x 1


You can customise

Decipher is easier



Links to OMIM