DataIntensive Scalability in Machine Learning and Computational Proteomics
Data-Intensive Scalability in Machine Learning and Computational Proteomics Jaime Carbonell et al Carnegie Mellon University jgc@cs. cmu. edu January, 2009
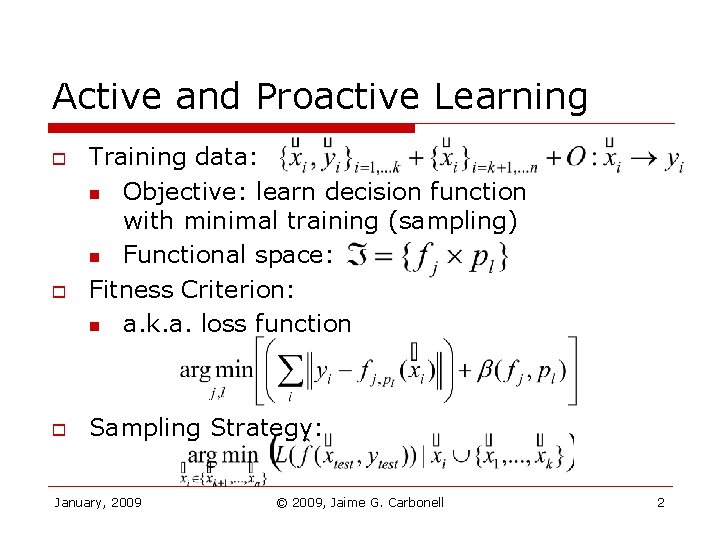
Active and Proactive Learning o o o Training data: n Objective: learn decision function with minimal training (sampling) n Functional space: Fitness Criterion: n a. k. a. loss function Sampling Strategy: January, 2009 © 2009, Jaime G. Carbonell 2
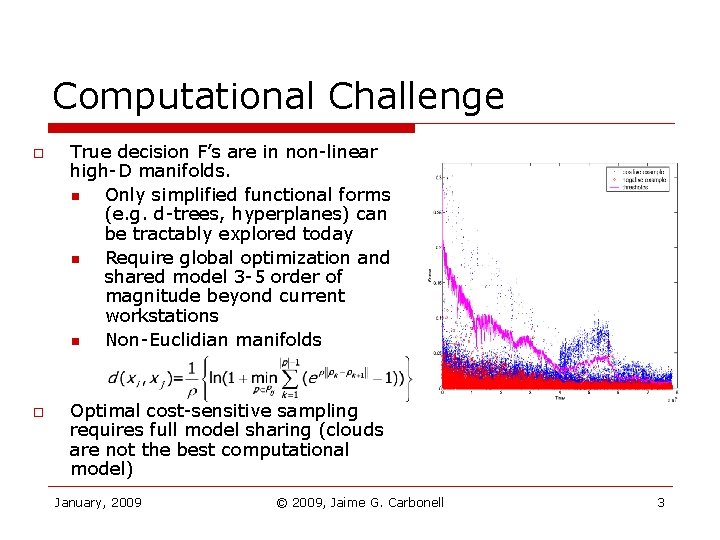
Computational Challenge o o True decision F’s are in non-linear high-D manifolds. n Only simplified functional forms (e. g. d-trees, hyperplanes) can be tractably explored today n Require global optimization and shared model 3 -5 order of magnitude beyond current workstations n Non-Euclidian manifolds Optimal cost-sensitive sampling requires full model sharing (clouds are not the best computational model) January, 2009 © 2009, Jaime G. Carbonell 3
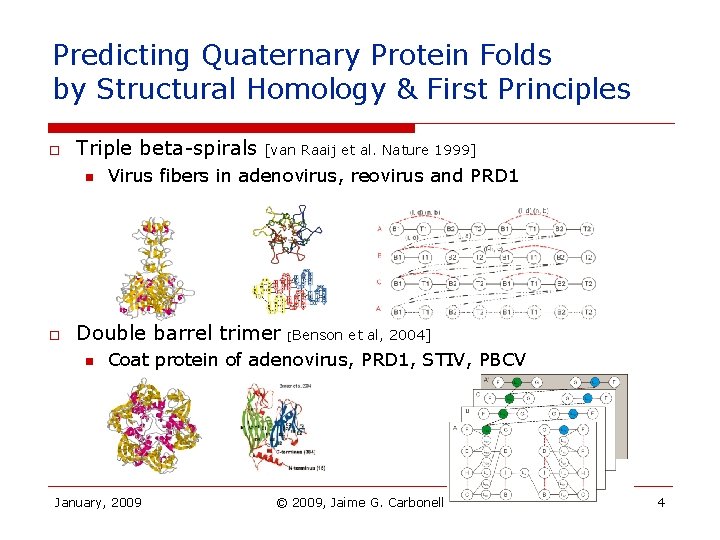
Predicting Quaternary Protein Folds by Structural Homology & First Principles o Triple beta-spirals n o [van Raaij et al. Nature 1999] Virus fibers in adenovirus, reovirus and PRD 1 Double barrel trimer n [Benson et al, 2004] Coat protein of adenovirus, PRD 1, STIV, PBCV January, 2009 © 2009, Jaime G. Carbonell 4
![Linked Segmentation Conditional Random Fields [Liu & Carbonell] o o Goal: Predict how protein Linked Segmentation Conditional Random Fields [Liu & Carbonell] o o Goal: Predict how protein](http://slidetodoc.com/presentation_image_h2/8b4f3e96f95c5d2762f22d14698f65e1/image-5.jpg)
Linked Segmentation Conditional Random Fields [Liu & Carbonell] o o Goal: Predict how protein complex will fold Nodes: Secondary protein structures and/or simple folds Edges: Local interactions and long-range inter-chain and intra -chain interactions L-SCRF: conditional probability of y given x is defined as Joint Labels January, 2009 © 2009, Jaime G. Carbonell 5
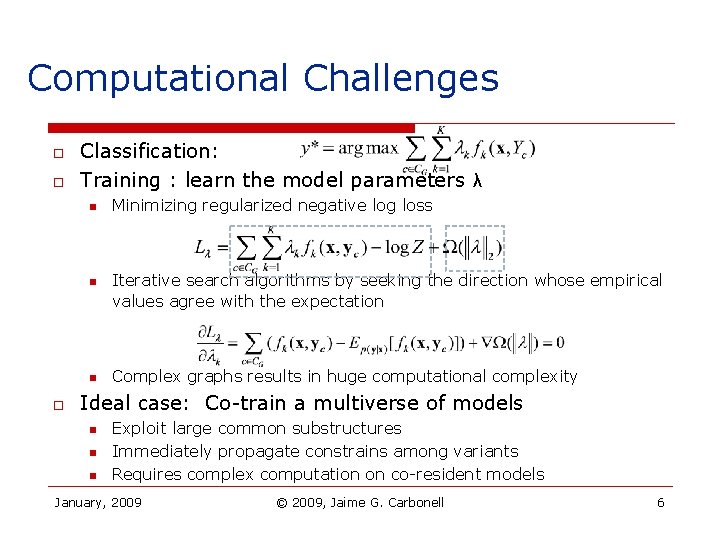
Computational Challenges o o Classification: Training : learn the model parameters λ n n n o Minimizing regularized negative log loss Iterative search algorithms by seeking the direction whose empirical values agree with the expectation Complex graphs results in huge computational complexity Ideal case: Co-train a multiverse of models n n n Exploit large common substructures Immediately propagate constrains among variants Requires complex computation on co-resident models January, 2009 © 2009, Jaime G. Carbonell 6
![Learning Protein Interaction Networks Intra- and Inter-Organism [Qi, Klein. Seetharaman, Tastan, Carbonell] PSB 05 Learning Protein Interaction Networks Intra- and Inter-Organism [Qi, Klein. Seetharaman, Tastan, Carbonell] PSB 05](http://slidetodoc.com/presentation_image_h2/8b4f3e96f95c5d2762f22d14698f65e1/image-7.jpg)
Learning Protein Interaction Networks Intra- and Inter-Organism [Qi, Klein. Seetharaman, Tastan, Carbonell] PSB 05 PROTEINS 06 BMC Bioinfo 07 CCR 08 Protein Complex ISMB 08 Pathway Pairwise Interactions PPI Network Human-PPI (Revise 08) HIV-Human PPI (Revise) Domain/Motif Interactions January, 2009 (Preparation) Function Func A Func ? Implication Genome Biology 08 © 2009, Jaime G. Carbonell 7
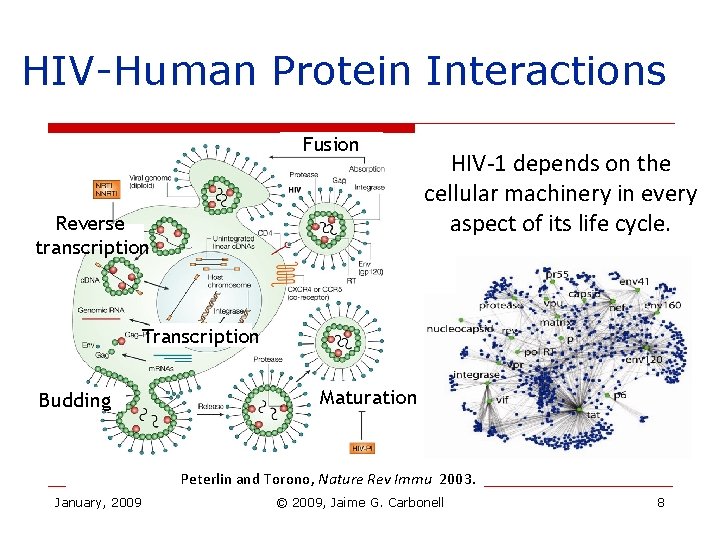
HIV-Human Protein Interactions Fusion Reverse transcription HIV-1 depends on the cellular machinery in every aspect of its life cycle. Transcription Budding Maturation Peterlin and Torono, Nature Rev Immu 2003. January, 2009 © 2009, Jaime G. Carbonell 8
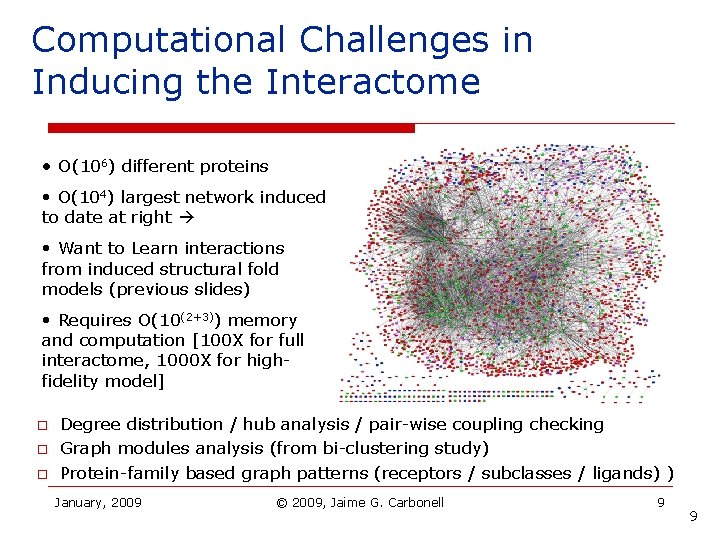
Computational Challenges in Inducing the Interactome • O(106) different proteins • O(104) largest network induced to date at right • Want to Learn interactions from induced structural fold models (previous slides) • Requires O(10(2+3)) memory and computation [100 X for full interactome, 1000 X for highfidelity model] o o o Degree distribution / hub analysis / pair-wise coupling checking Graph modules analysis (from bi-clustering study) Protein-family based graph patterns (receptors / subclasses / ligands) ) January, 2009 © 2009, Jaime G. Carbonell 9 9
- Slides: 9