Data Type 1 Microarrays Reverse Genetics approach Genomics












- Slides: 23

Data Type 1: Microarrays • Reverse Genetics approach • Genomics • So we need to understand what exactly is a Microarray • DNA Microarrays are small, solid supports(the size of two side -by-side pinky fingers) onto which the sequences from thousands of different genes are immobilized, or attached, at fixed locations. • So we need to KNOW the sequence to design this array.

Definition of Microarray A semiconductor device that is used to detect the DNA makeup of a cell. • It contains hundreds of thousands of tiny squares designed to mate with a particular gene. • They react to the liquified cells poured over it and are detectable by a laser. • Microarrays were thought of the way to get all answers for a while. • Sometimes called "biochips, " micro arrays are commonly known as "gene chips, " Gene. Chip is an Affymetrix trademark.

Microarrays have • revealed new patterns of coordinated gene expression across gene families, • expanded the size of existing gene families, • increased our understanding of how these genes coordinate • precise knowledge of these inter-relationships has emerged • speeded the identification of genes involved in the development of various diseases • aided the examination of the integration of gene expression and function at the cell level, • revealed how multiple gene products work together

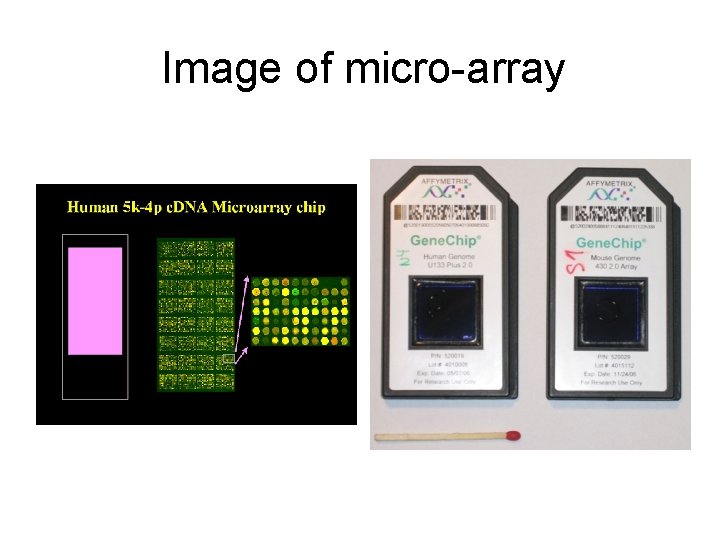
Image of micro-array

Types of Microarray • • Gene Expression Comparative Hybridization Gene ID Ch. Ip SNP detection Alternate Splicing Tiling Arrays • In the first part of the class we will focus on Gene Expression and the later part look at SNP detection.

Gene Expression Analysis • Typical Northern Blot: One gene/ experiment/ more than one sample – Fairly quantitative – Time consuming – Limited information • Microarray and RNA-seq: thousands of gene/one sample – Fairly quantitative – Less time – Massive information

REMEMBER: Central dogma of molecular biology • Each gene is transcribed (at the appropriate time) from DNA into m. RNA, which then leaves the nucleus and is translated into the required protein. • This is the principle used for microarrays

Simple Cartoon of the idea of Microarray(borrowed from Bumgarner 2104) Array surface A B Solution A A A B A B After hybridization B

Hybridization

AFFY chip for gene expression profiling There are many others but we will focus on Affy

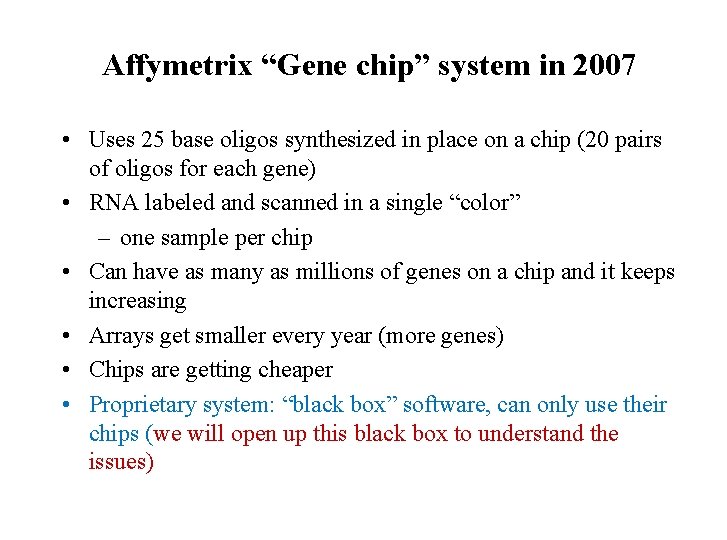
Affymetrix “Gene chip” system in 2007 • Uses 25 base oligos synthesized in place on a chip (20 pairs of oligos for each gene) • RNA labeled and scanned in a single “color” – one sample per chip • Can have as many as millions of genes on a chip and it keeps increasing • Arrays get smaller every year (more genes) • Chips are getting cheaper • Proprietary system: “black box” software, can only use their chips (we will open up this black box to understand the issues)

A bit about the technology • • Affy is an "oligonucleotide array“- referring to the production method produced by printing short oligonucleotide sequences designed to represent a single gene/family/splice-variants Direct synthesizing this sequence rather than depositing whole intact sequences. Sequences may be longer (60 -mer probes such as the Agilent design) or shorter (25 -mer probes produced by Affymetrix) depending on the desired purpose;

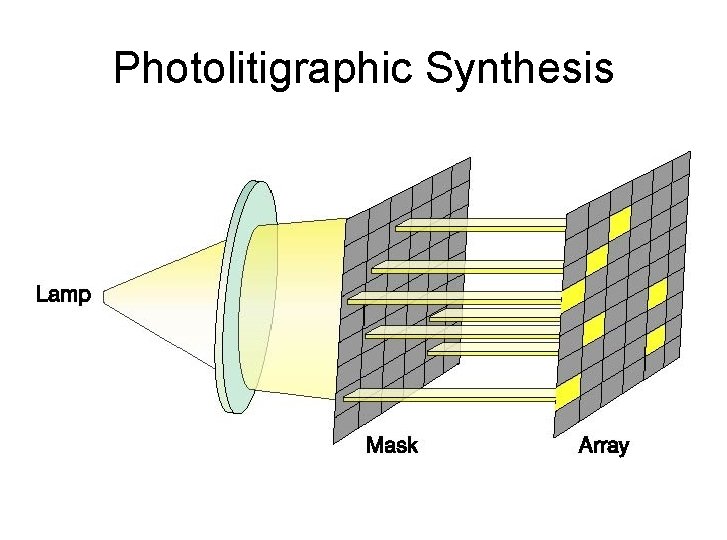
Technology contd… • longer probes to target individual genes, • shorter probes may be spotted in higher density across the array are cheaper • include photolithographic synthesis (Affymetrix) on a silica substrate where light and light-sensitive masking agents are used to "build" a sequence one nucleotide at a time across the entire array. • Each applicable probe is selectively "unmasked“. Done at a time. • After many repetitions, the sequences of every probe become fully constructed. • The problem of non-specific hybridization always exists • Uses perfect match and mismatch probes to look at non-specific hybridization.


Problem of Non-specific binding • Similar genes can bind on the probes that are not the correct gene of interest. • This is why Affy uses the mis-match idea


Probe Design: Affymetrix

Photolitigraphic Synthesis

Scanning AFFY chips • Light removes protecting groups at defined positions. • Single nucleotide washed over the chip, binds where the protecting group removed. • Through successive steps, any sequence can be built up in any position on the chip. • The number of steps corresponds with length of oligo, so can increase # of genes without # of steps

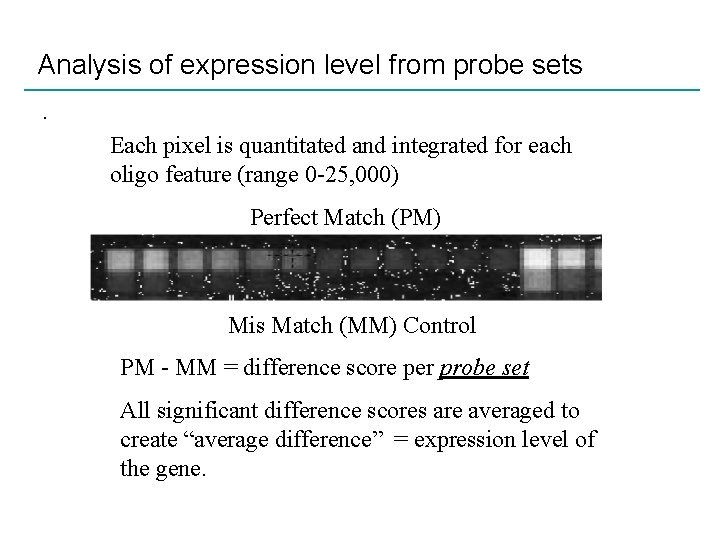
Analysis of expression level from probe sets. Each pixel is quantitated and integrated for each oligo feature (range 0 -25, 000) Perfect Match (PM) Mis Match (MM) Control PM - MM = difference score per probe set All significant difference scores are averaged to create “average difference” = expression level of the gene.

Microarray Data Analysis How to Handle Microarray Data? Preprocessing: • Signal Generation from Image • Normalization • Filtering Analysis: • Statistical Tests for differential Expressions, t tests, non-parametric Tests, ANOVA • Clustering: Hierarchical, non-hierarchichal, SOM • Classification: Discriminant Analysis, PCA The Main Goal of Microarray Data Analysis is to Generate a List Of ‘Interesting’ Genes

Current Affymetrix • Many products with lot more options for custom arrays. • Have products that can handle multiple samples at once. • Description for Affymetrix HG-U 133, MG-430, and RG-230 Array • These data sources are used to design probes that interrogate 9 to 11 unique sequences of each transcript. • The unique 25 -mer probes interrogate up to 275 bases per transcript.

Limitations of Microarrays • They are an INDRECT measure of relative concentration • The signal measured is ASSUMED to be proportional to the species concentration • At high concentrations we can have saturation • At low concentrations – no binding • A probe might be designed for gene A, but genes B, C and D can incorrectly bind if they have similar homology • Can only detect KNOWN sequences