Data Mining Techniques For Correlating Phenotypic Expressions With
- Slides: 1

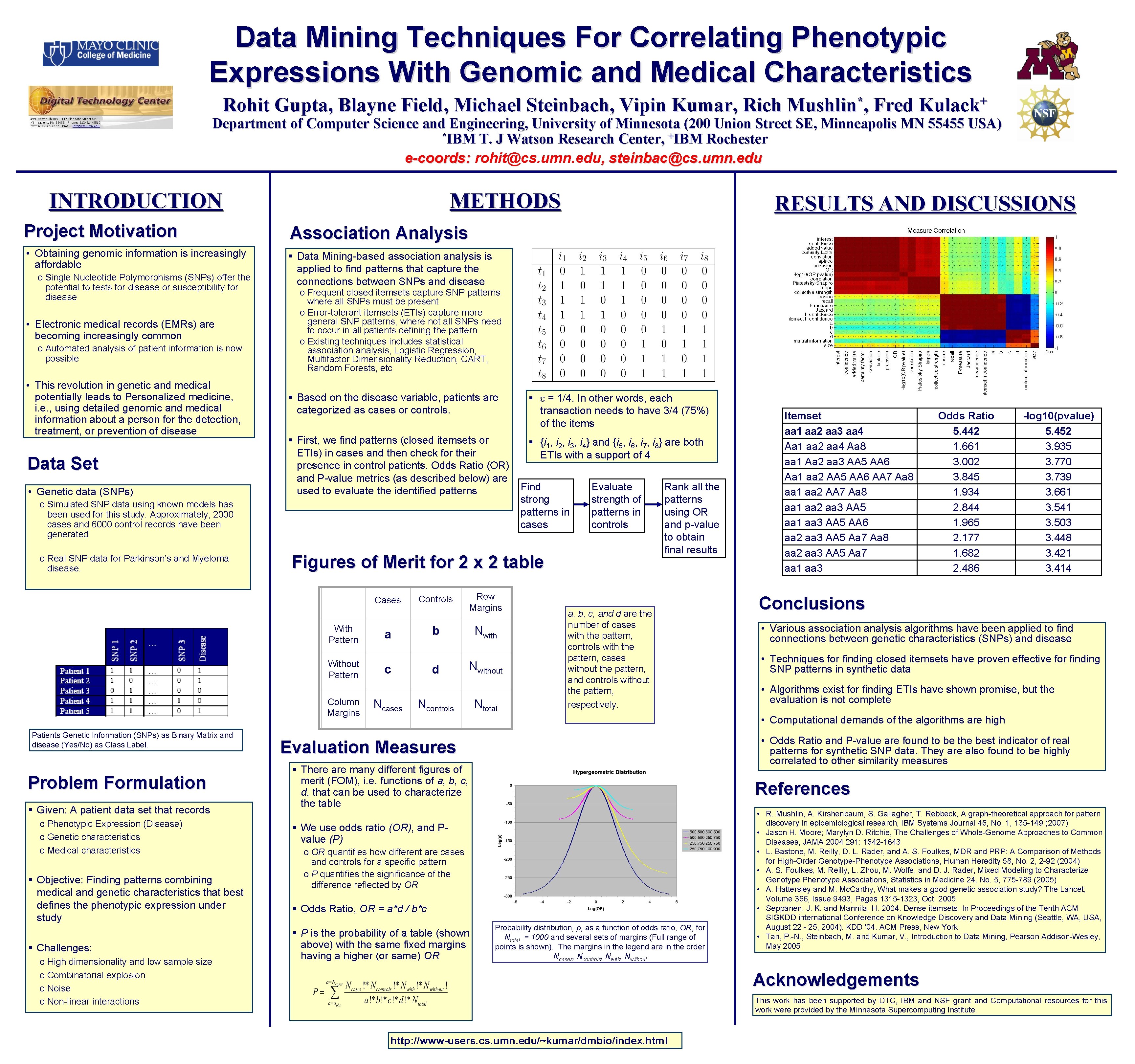
Data Mining Techniques For Correlating Phenotypic Expressions With Genomic and Medical Characteristics Rohit Gupta, Blayne Field, Michael Steinbach, Vipin Kumar, Rich * Mushlin , Fred + Kulack Department of Computer Science and Engineering, University of Minnesota (200 Union Street SE, Minneapolis MN 55455 USA) *IBM T. J Watson Research Center, +IBM Rochester e-coords: rohit@cs. umn. edu, steinbac@cs. umn. edu INTRODUCTION METHODS Project Motivation Association Analysis • Obtaining genomic information is increasingly affordable § Data Mining-based association analysis is applied to find patterns that capture the connections between SNPs and disease o Single Nucleotide Polymorphisms (SNPs) offer the potential to tests for disease or susceptibility for disease • Electronic medical records (EMRs) are becoming increasingly common o Automated analysis of patient information is now possible • This revolution in genetic and medical potentially leads to Personalized medicine, i. e. , using detailed genomic and medical information about a person for the detection, treatment, or prevention of disease Data Set • Genetic data (SNPs) o Frequent closed itemsets capture SNP patterns where all SNPs must be present o Error-tolerant itemsets (ETIs) capture more general SNP patterns, where not all SNPs need to occur in all patients defining the pattern o Existing techniques includes statistical association analysis, Logistic Regression, Multifactor Dimensionality Reduction, CART, Random Forests, etc § Based on the disease variable, patients are categorized as cases or controls. § = 1/4. In other words, each transaction needs to have 3/4 (75%) of the items § First, we find patterns (closed itemsets or ETIs) in cases and then check for their presence in control patients. Odds Ratio (OR) and P-value metrics (as described below) are used to evaluate the identified patterns § {i 1, i 2, i 3, i 4} and {i 5, i 6, i 7, i 8} are both ETIs with a support of 4 o Simulated SNP data using known models has been used for this study. Approximately, 2000 cases and 6000 control records have been generated o Real SNP data for Parkinson’s and Myeloma disease. With Pattern Without Pattern Column Margins Problem Formulation § Given: A patient data set that records o Phenotypic Expression (Disease) o Genetic characteristics o Medical characteristics § Objective: Finding patterns combining medical and genetic characteristics that best defines the phenotypic expression under study § Challenges: o High dimensionality and low sample size o Combinatorial explosion o Noise o Non-linear interactions Find strong patterns in cases Evaluate strength of patterns in controls Figures of Merit for 2 x 2 table Cases Patients Genetic Information (SNPs) as Binary Matrix and disease (Yes/No) as Class Label. RESULTS AND DISCUSSIONS Controls b a c Ncases Row Margins d Nwithout Ncontrols Ntotal Rank all the patterns using OR and p-value to obtain final results a, b, c, and d are the number of cases with the pattern, controls with the pattern, cases without the pattern, and controls without the pattern, respectively. Itemset aa 1 aa 2 aa 3 aa 4 Aa 1 aa 2 aa 4 Aa 8 aa 1 Aa 2 aa 3 AA 5 AA 6 Aa 1 aa 2 AA 5 AA 6 AA 7 Aa 8 aa 1 aa 2 aa 3 AA 5 aa 1 aa 3 AA 5 AA 6 aa 2 aa 3 AA 5 Aa 7 Aa 8 aa 2 aa 3 AA 5 Aa 7 aa 1 aa 3 Odds Ratio 5. 442 1. 661 3. 002 3. 845 1. 934 2. 844 1. 965 2. 177 1. 682 2. 486 -log 10(pvalue) 5. 452 3. 935 3. 770 3. 739 3. 661 3. 541 3. 503 3. 448 3. 421 3. 414 Conclusions • Various association analysis algorithms have been applied to find connections between genetic characteristics (SNPs) and disease • Techniques for finding closed itemsets have proven effective for finding SNP patterns in synthetic data • Algorithms exist for finding ETIs have shown promise, but the evaluation is not complete • Computational demands of the algorithms are high • Odds Ratio and P-value are found to be the best indicator of real patterns for synthetic SNP data. They are also found to be highly correlated to other similarity measures Evaluation Measures § There are many different figures of merit (FOM), i. e. functions of a, b, c, d, that can be used to characterize the table References § We use odds ratio (OR), and Pvalue (P) o OR quantifies how different are cases and controls for a specific pattern o P quantifies the significance of the difference reflected by OR § Odds Ratio, OR = a*d / b*c § P is the probability of a table (shown above) with the same fixed margins having a higher (or same) OR Probability distribution, p, as a function of odds ratio, OR, for Ntotal = 1000 and several sets of margins (Full range of points is shown). The margins in the legend are in the order Ncases, Ncontrols, Nwithout • R. Mushlin, A. Kirshenbaum, S. Gallagher, T. Rebbeck, A graph-theoretical approach for pattern discovery in epidemiological research, IBM Systems Journal 46, No. 1, 135 -149 (2007) • Jason H. Moore; Marylyn D. Ritchie, The Challenges of Whole-Genome Approaches to Common Diseases, JAMA 2004 291: 1642 -1643 • L. Bastone, M. Reilly, D. L. Rader, and A. S. Foulkes, MDR and PRP: A Comparison of Methods for High-Order Genotype-Phenotype Associations, Human Heredity 58, No. 2, 2 -92 (2004) • A. S. Foulkes, M. Reilly, L. Zhou, M. Wolfe, and D. J. Rader, Mixed Modeling to Characterize Genotype Phenotype Associations, Statistics in Medicine 24, No. 5, 775 -789 (2005) • A. Hattersley and M. Mc. Carthy, What makes a good genetic association study? The Lancet, Volume 366, Issue 9493, Pages 1315 -1323, Oct. 2005 • Seppänen, J. K. and Mannila, H. 2004. Dense itemsets. In Proceedings of the Tenth ACM SIGKDD international Conference on Knowledge Discovery and Data Mining (Seattle, WA, USA, August 22 - 25, 2004). KDD '04. ACM Press, New York • Tan, P. -N. , Steinbach, M. and Kumar, V. , Introduction to Data Mining, Pearson Addison-Wesley, May 2005 Acknowledgements This work has been supported by DTC, IBM and NSF grant and Computational resources for this work were provided by the Minnesota Supercomputing Institute. http: //www-users. cs. umn. edu/~kumar/dmbio/index. html