Data Mining Principles and Algorithms Graph Pattern Mining




- Slides: 72

Data Mining: Principles and Algorithms Graph Pattern Mining Jiawei Han Department of Computer Science University of Illinois at Urbana-Champaign www. cs. uiuc. edu/~hanj © 2013 Jiawei Han. All rights reserved. 1


Graph Mining n Graph Pattern Mining Frequent Subgraph Patterns n Impact on Graph Search I: Graph Indexing n Impact on Graph Search II: Graph Similarity Search n Graph Classification (not to be covered) n Graph Clustering (not to be covered) n Summary 3

Why Graph Mining? n n Graphs are ubiquitous n Chemical compounds (Cheminformatics) n Protein structures, biological pathways/networks (Bioinformactics) n Program control flow, traffic flow, and workflow analysis n XML databases, Web, and social network analysis Graph is a general model n n Diversity of graphs n n Trees, lattices, sequences, and items are degenerated graphs Directed vs. undirected, labeled vs. unlabeled (edges & vertices), weighted, with angles & geometry (topological vs. 2 -D/3 -D) Complexity of algorithms: many problems are of high complexity 4

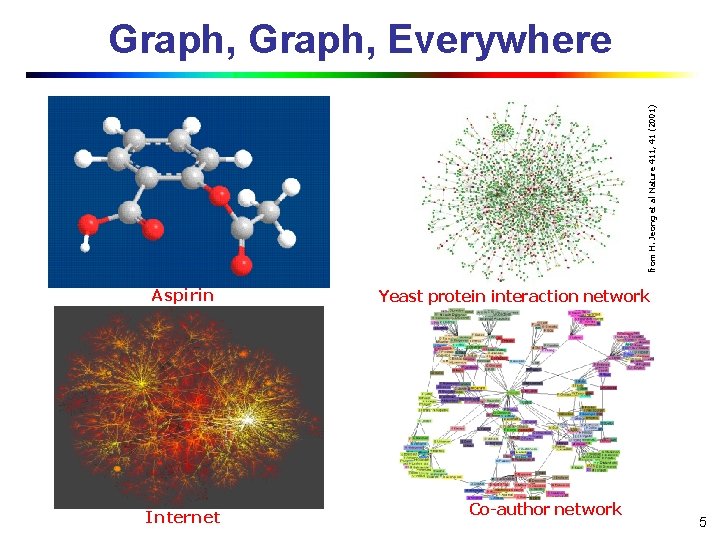
from H. Jeong et al Nature 411, 41 (2001) Graph, Everywhere Aspirin Internet Yeast protein interaction network Co-author network 5

Graph Pattern Mining n Frequent subgraphs n A (sub)graph is frequent if its support (occurrence frequency) in a given dataset is no less than a minimum support threshold n Applications of graph pattern mining n Mining biochemical structures n Program control flow analysis n Mining XML structures or Web communities n Building blocks for graph classification, clustering, compression, comparison, and correlation analysis 6

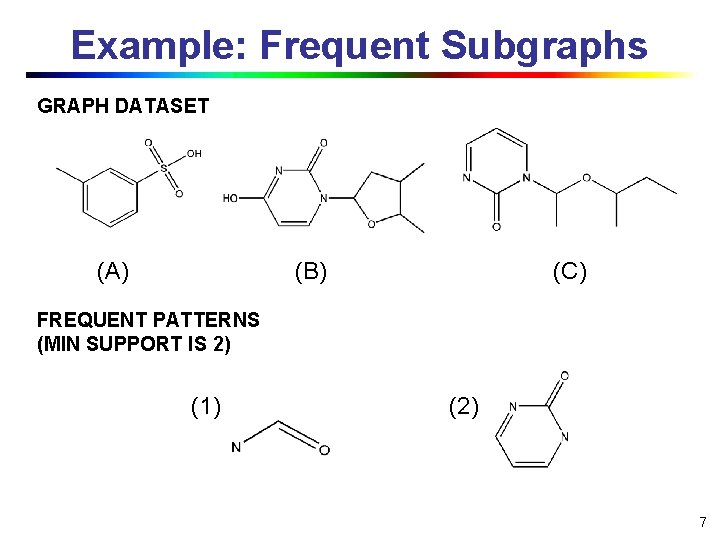
Example: Frequent Subgraphs GRAPH DATASET (A) (B) (C) FREQUENT PATTERNS (MIN SUPPORT IS 2) (1) (2) 7

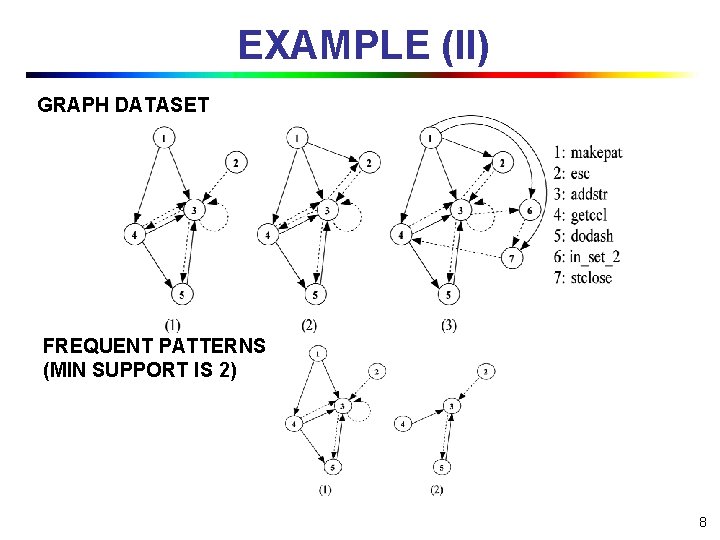
EXAMPLE (II) GRAPH DATASET FREQUENT PATTERNS (MIN SUPPORT IS 2) 8

Graph Mining Algorithms n Incomplete beam search – Greedy (Subdue) n Inductive logic programming (WARMR) n Graph theory-based approaches n Apriori-based approach n Pattern-growth approach 9

SUBDUE (Holder et al. KDD’ 94) n Start with single vertices n Expand best substructures with a new edge n Limit the number of best substructures n n Substructures are evaluated based on their ability to compress input graphs Using minimum description length (DL) Best substructure S in graph G minimizes: DL(S) + DL(GS) Terminate until no new substructure is discovered 10

WARMR (Dehaspe et al. KDD’ 98) n Graphs are represented by Datalog facts n atomel(C, A 1, c), bond (C, A 1, A 2, BT), atomel(C, A 2, c) : a carbon atom bound to a carbon atom with bond type BT n WARMR: the first general purpose ILP system n Level-wise search n Simulate Apriori for frequent pattern discovery 11

Frequent Subgraph Mining Approaches n n Apriori-based approach n AGM/Ac. GM: Inokuchi, et al. (PKDD’ 00) n FSG: Kuramochi and Karypis (ICDM’ 01) # n PATH : Vanetik and Gudes (ICDM’ 02, ICDM’ 04) n FFSM: Huan, et al. (ICDM’ 03) Pattern growth approach n Mo. Fa, Borgelt and Berthold (ICDM’ 02) n g. Span: Yan and Han (ICDM’ 02) n Gaston: Nijssen and Kok (KDD’ 04) 12

Properties of Graph Mining Algorithms n n n Search order n breadth vs. depth Generation of candidate subgraphs n apriori vs. pattern growth Elimination of duplicate subgraphs n passive vs. active Support calculation n embedding store or not Discover order of patterns n path tree graph 13

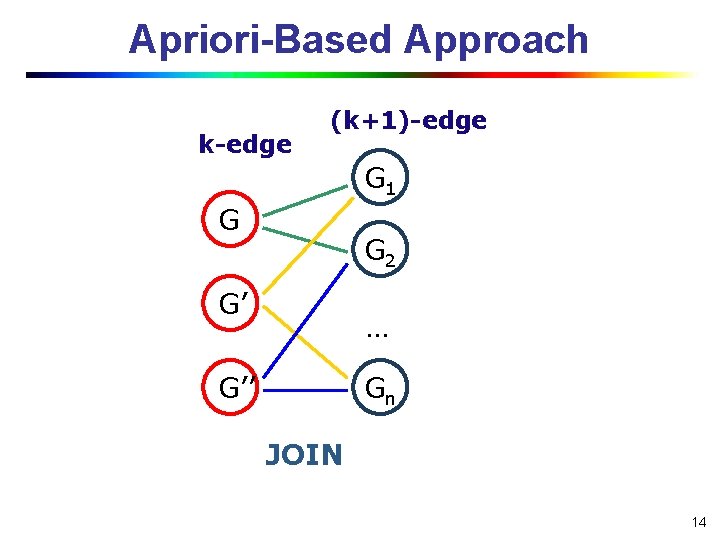
Apriori-Based Approach k-edge (k+1)-edge G 1 G G 2 G’ … G’’ Gn JOIN 14

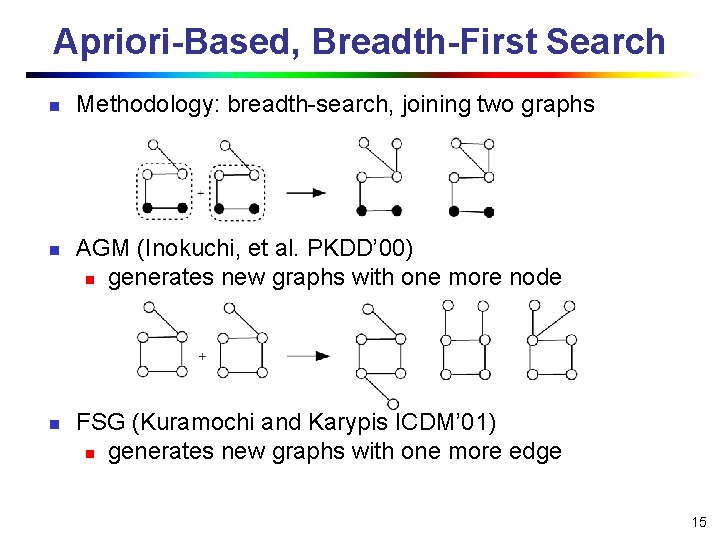
Apriori-Based, Breadth-First Search n n n Methodology: breadth-search, joining two graphs AGM (Inokuchi, et al. PKDD’ 00) n generates new graphs with one more node FSG (Kuramochi and Karypis ICDM’ 01) n generates new graphs with one more edge 15

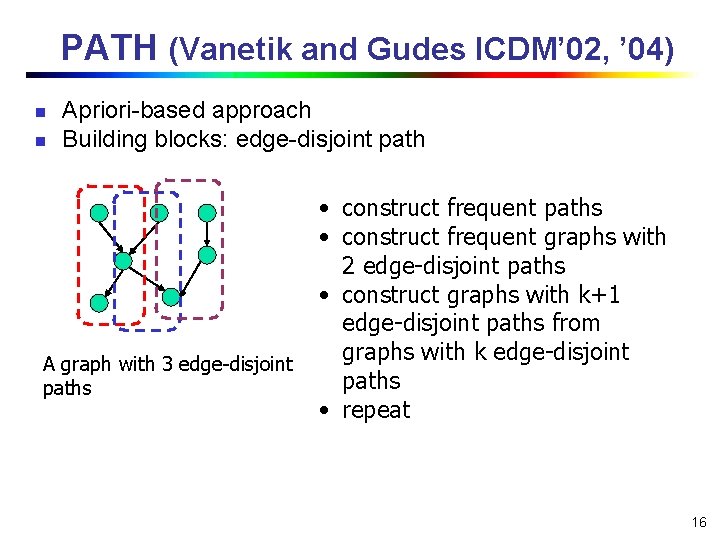
PATH (Vanetik and Gudes ICDM’ 02, ’ 04) n n Apriori-based approach Building blocks: edge-disjoint path A graph with 3 edge-disjoint paths • construct frequent paths • construct frequent graphs with 2 edge-disjoint paths • construct graphs with k+1 edge-disjoint paths from graphs with k edge-disjoint paths • repeat 16

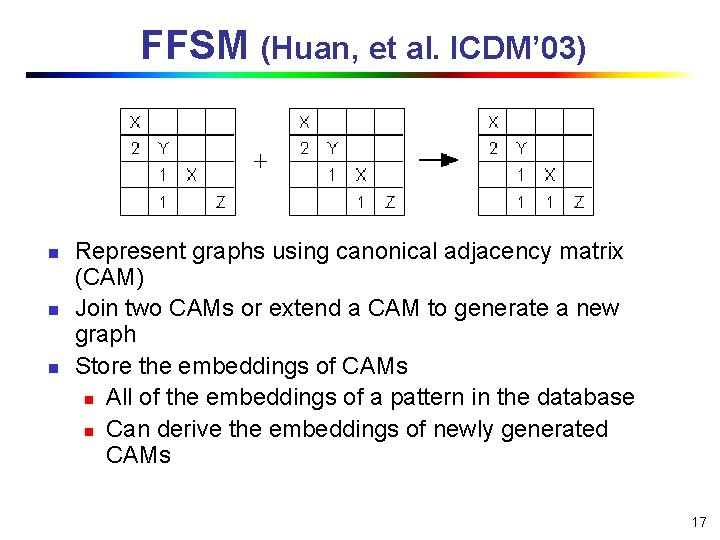
FFSM (Huan, et al. ICDM’ 03) n n n Represent graphs using canonical adjacency matrix (CAM) Join two CAMs or extend a CAM to generate a new graph Store the embeddings of CAMs n All of the embeddings of a pattern in the database n Can derive the embeddings of newly generated CAMs 17

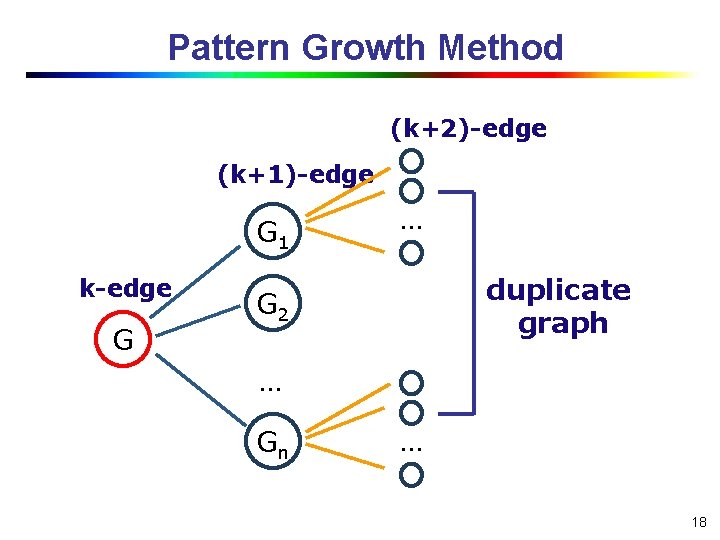
Pattern Growth Method (k+2)-edge (k+1)-edge G 1 k-edge G … duplicate graph G 2 … Gn … 18

Mo. Fa (Borgelt and Berthold ICDM’ 02) n Extend graphs by adding a new edge n Store embeddings of discovered frequent graphs n n Fast support calculation Also used in other later developed algorithms such as FFSM and GASTON Expensive Memory usage Local structural pruning n n 19

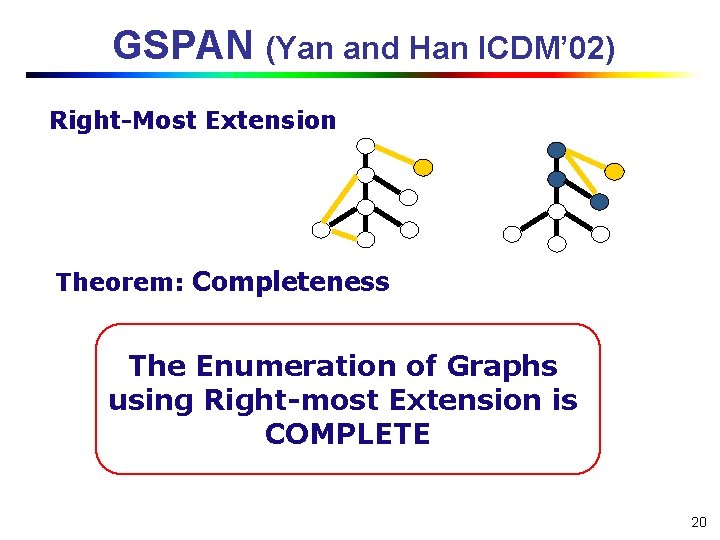
GSPAN (Yan and Han ICDM’ 02) Right-Most Extension Theorem: Completeness The Enumeration of Graphs using Right-most Extension is COMPLETE 20

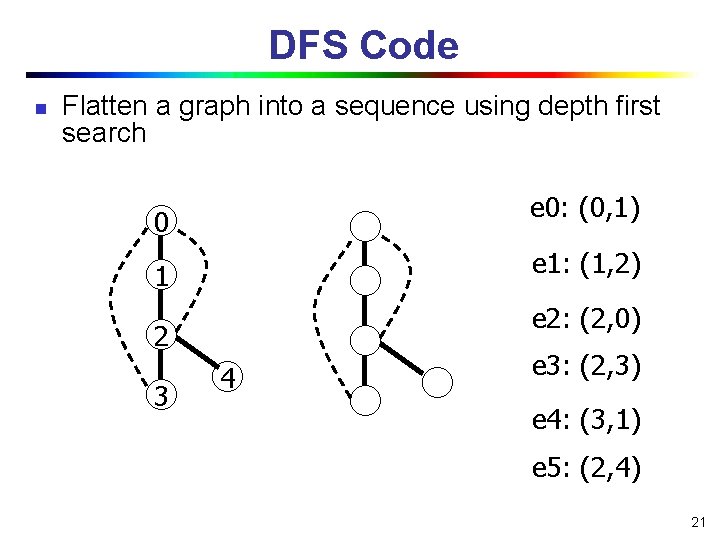
DFS Code n Flatten a graph into a sequence using depth first search e 0: (0, 1) 0 e 1: (1, 2) 1 e 2: (2, 0) 2 3 4 e 3: (2, 3) e 4: (3, 1) e 5: (2, 4) 21

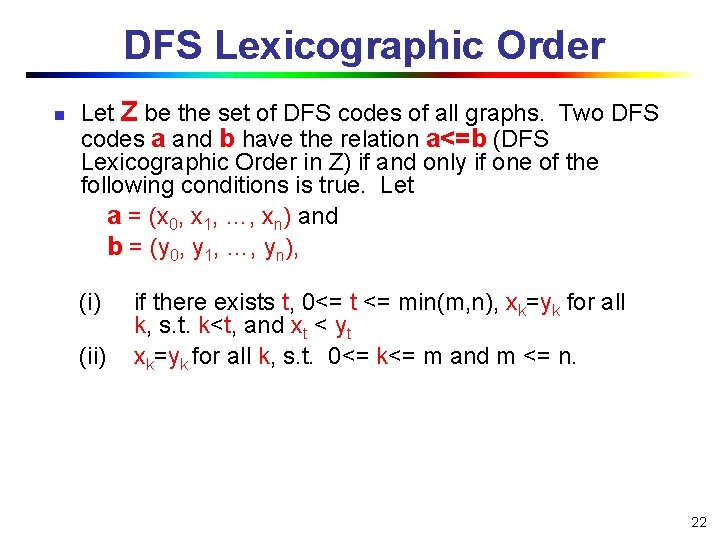
DFS Lexicographic Order n Let Z be the set of DFS codes of all graphs. Two DFS codes a and b have the relation a<=b (DFS Lexicographic Order in Z) if and only if one of the following conditions is true. Let a = (x 0, x 1, …, xn) and b = (y 0, y 1, …, yn), (i) (ii) if there exists t, 0<= t <= min(m, n), xk=yk for all k, s. t. k<t, and xt < yt xk=yk for all k, s. t. 0<= k<= m and m <= n. 22

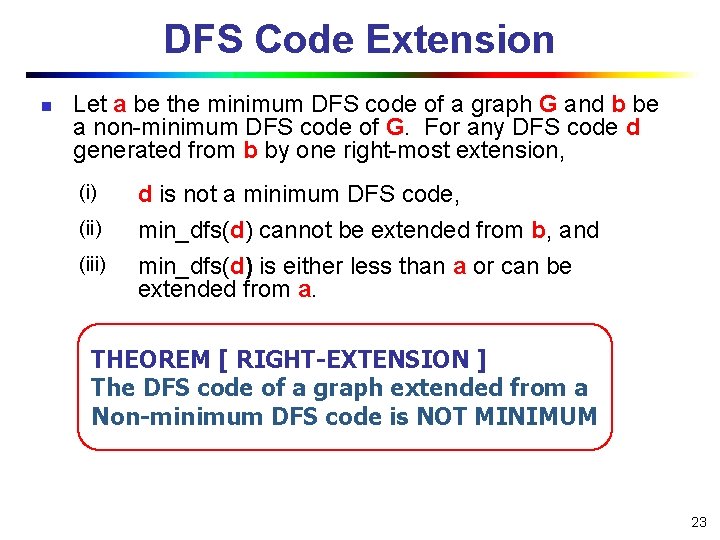
DFS Code Extension n Let a be the minimum DFS code of a graph G and b be a non-minimum DFS code of G. For any DFS code d generated from b by one right-most extension, (i) (iii) d is not a minimum DFS code, min_dfs(d) cannot be extended from b, and min_dfs(d) is either less than a or can be extended from a. THEOREM [ RIGHT-EXTENSION ] The DFS code of a graph extended from a Non-minimum DFS code is NOT MINIMUM 23

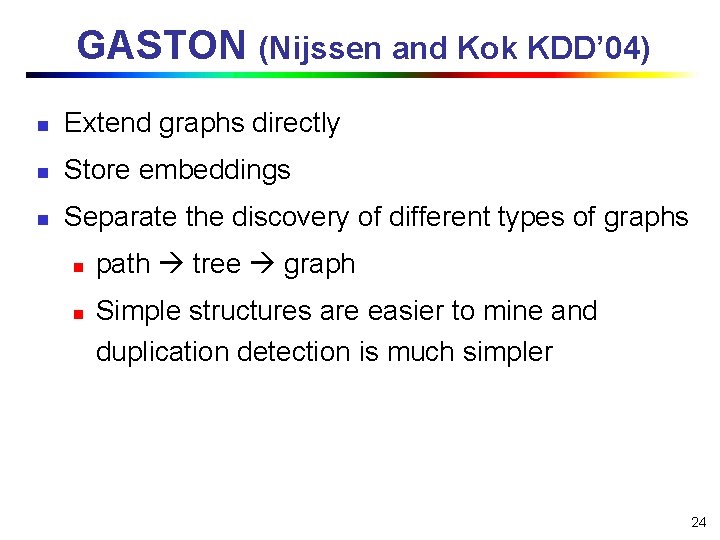
GASTON (Nijssen and Kok KDD’ 04) n Extend graphs directly n Store embeddings n Separate the discovery of different types of graphs n n path tree graph Simple structures are easier to mine and duplication detection is much simpler 24

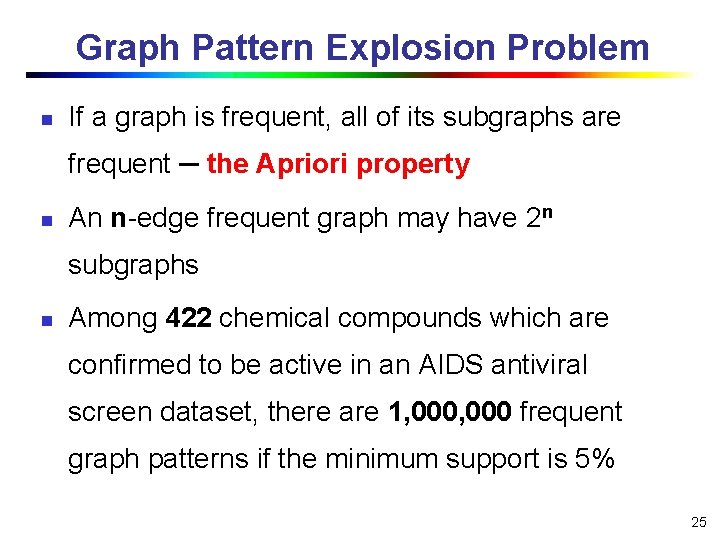
Graph Pattern Explosion Problem n If a graph is frequent, all of its subgraphs are frequent ─ the Apriori property n An n-edge frequent graph may have 2 n subgraphs n Among 422 chemical compounds which are confirmed to be active in an AIDS antiviral screen dataset, there are 1, 000 frequent graph patterns if the minimum support is 5% 25

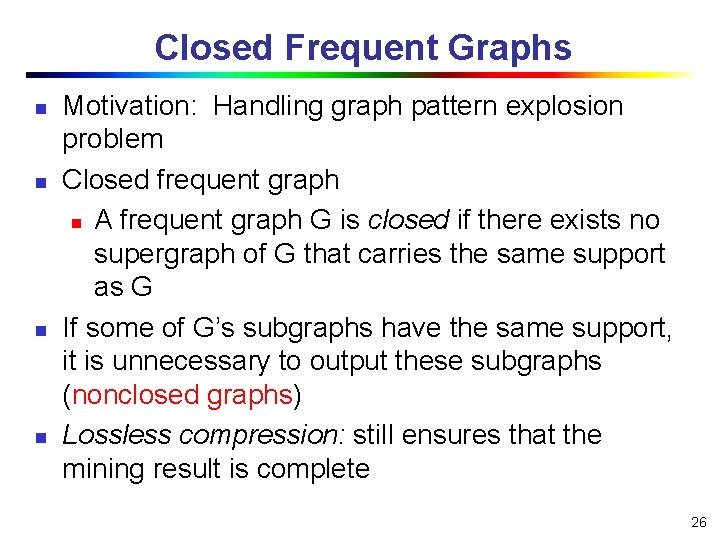
Closed Frequent Graphs n n Motivation: Handling graph pattern explosion problem Closed frequent graph n A frequent graph G is closed if there exists no supergraph of G that carries the same support as G If some of G’s subgraphs have the same support, it is unnecessary to output these subgraphs (nonclosed graphs) Lossless compression: still ensures that the mining result is complete 26

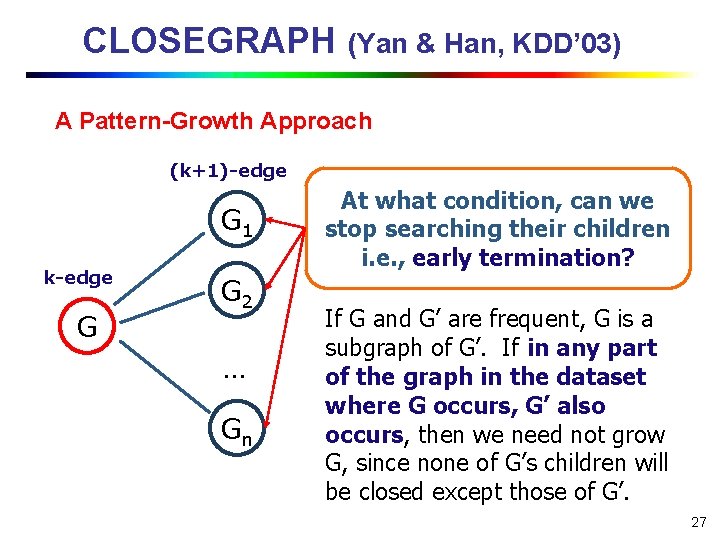
CLOSEGRAPH (Yan & Han, KDD’ 03) A Pattern-Growth Approach (k+1)-edge G 1 k-edge G G 2 … Gn At what condition, can we stop searching their children i. e. , early termination? If G and G’ are frequent, G is a subgraph of G’. If in any part of the graph in the dataset where G occurs, G’ also occurs, then we need not grow G, since none of G’s children will be closed except those of G’. 27

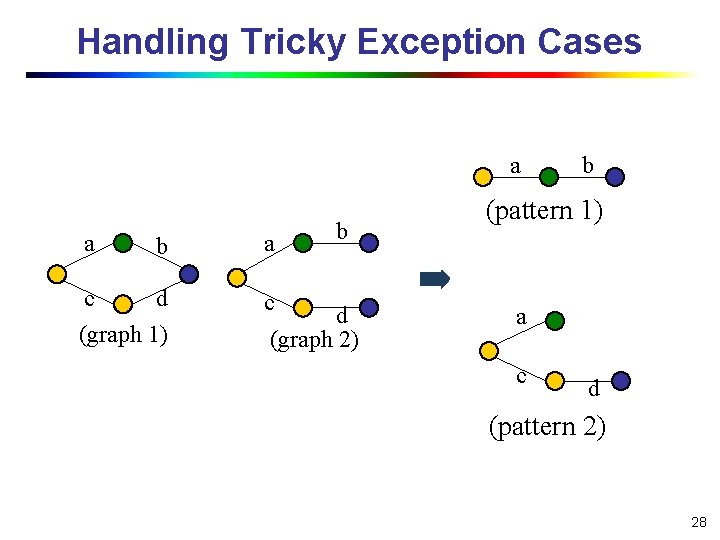
Handling Tricky Exception Cases a a b a c d (graph 1) c b d (graph 2) b (pattern 1) a c d (pattern 2) 28

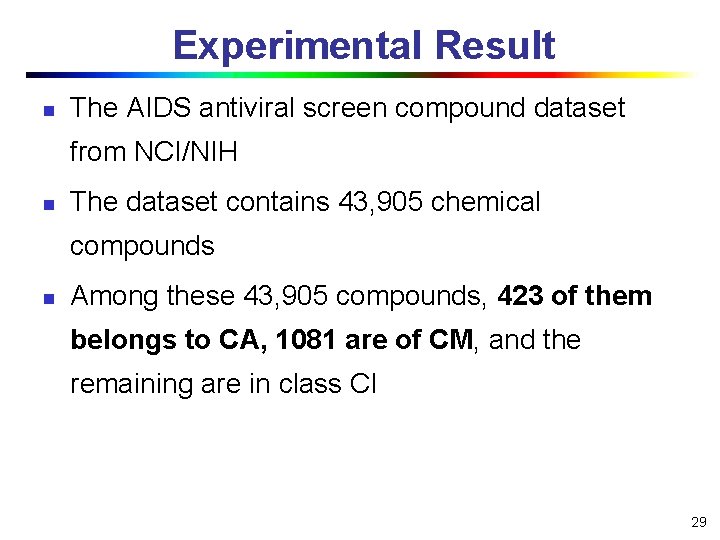
Experimental Result n The AIDS antiviral screen compound dataset from NCI/NIH n The dataset contains 43, 905 chemical compounds n Among these 43, 905 compounds, 423 of them belongs to CA, 1081 are of CM, and the remaining are in class CI 29

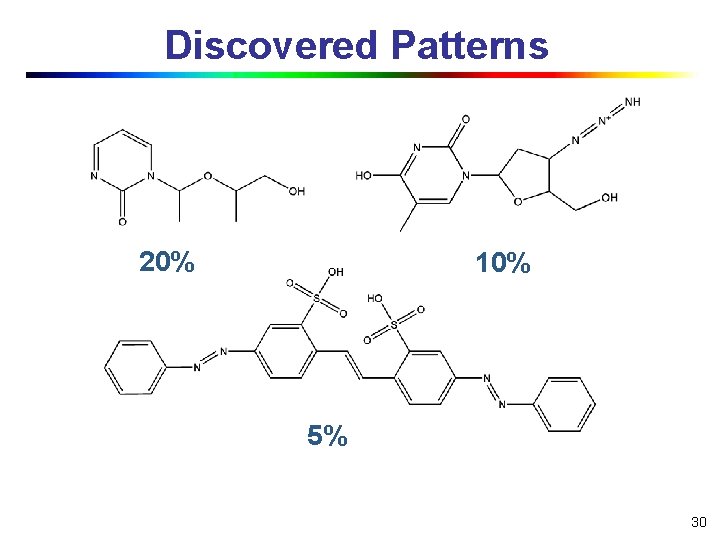
Discovered Patterns 20% 10% 5% 30

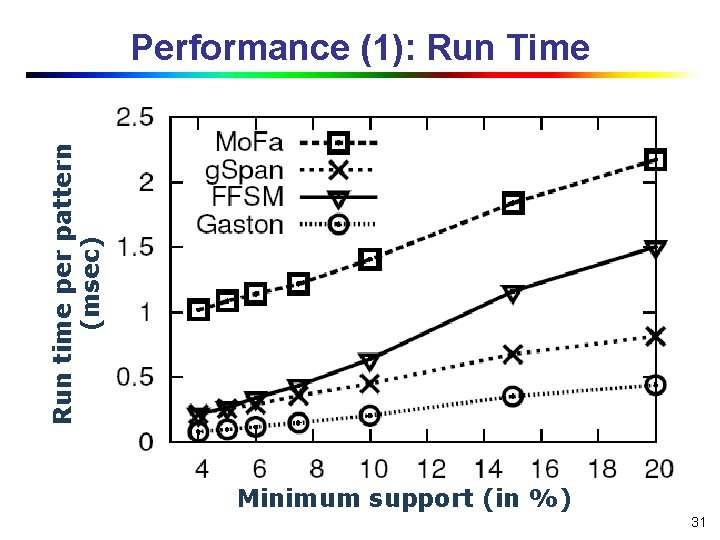
Run time per pattern (msec) Performance (1): Run Time Minimum support (in %) 31

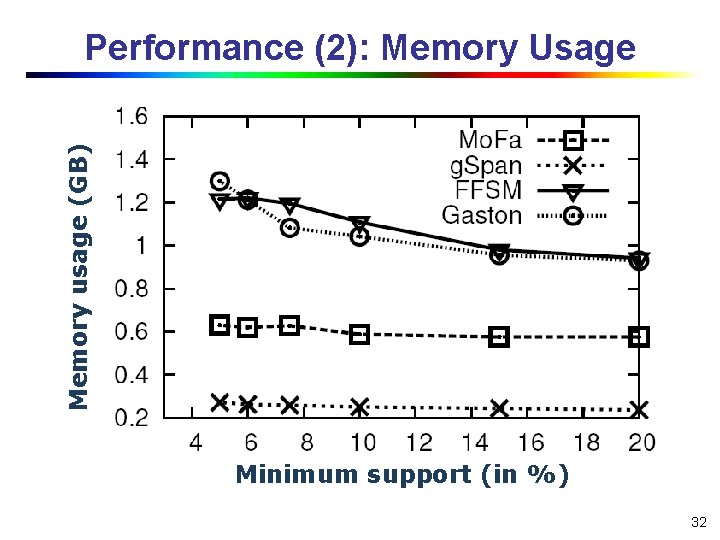
Memory usage (GB) Performance (2): Memory Usage Minimum support (in %) 32

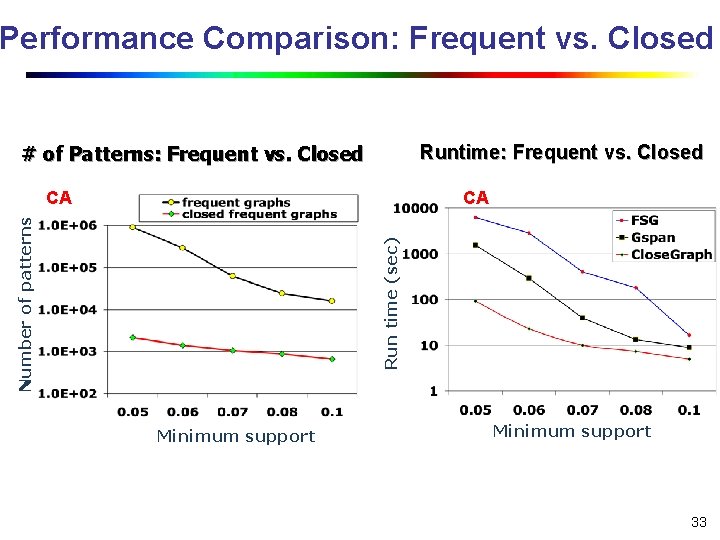
Performance Comparison: Frequent vs. Closed Runtime: Frequent vs. Closed # of Patterns: Frequent vs. Closed CA Run time (sec) Number of patterns CA Minimum support 33

Do the Odds Beat the Curse of Complexity? n n Potentially exponential number of frequent patterns n The worst case complexty vs. the expected probability 4 n Ex. : Suppose Walmart has 10 kinds of products -4 n The chance to pick up one product 10 -40 n The chance to pick up a particular set of 10 products: 10 n What is the chance this particular set of 10 products to be frequent 103 times in 109 transactions? Have we solved the NP-hard problem of subgraph isomorphism testing? n No. But the real graphs in bio/chemistry is not so bad n A carbon has only 4 bounds and most proteins in a network have distinct labels 34

Graph Mining n Graph Pattern Mining Frequent Subgraph Patterns n Impact on Graph Search I: Graph Indexing n Impact on Graph Search II: d Graph Similarity e r e ov C m a e x b Search E t to rm n er T No d Mi e h Graph Classification (not to be covered) in t n Graph Clustering (not to be covered) n Summary 35

Graph Search n Querying graph databases: n Given a graph database and a query graph, find all the graphs containing this query graph ed r e v Co m a e x b E to t rm o e T N id M e in th query graph database 36

Scalability Issue n n Sequential scan n Disk I/Os n Subgraph isomorphism testing An indexing mechanism is needed n n n ed r e v o Day. Light: Daylight. com C(commercial) m a e x b E to t rm o e T N Graph. Grep: Dennis. MShasha, et al. PODS'02 id e th n i Grace: Srinath Srinivasa, et al. ICDE'03 37

Indexing Strategy Query graph (Q) Graph (G) If graph G contains query graph Q, G should contain any substructure of Q ed r e v Co m a e x b E to t rm o e T Substructure N id M e in th Remarks n Index substructures of a query graph to prune graphs that do not contain these substructures 38

Indexing Framework n Two steps in processing graph queries Step 1. Index Construction n Enumerate structures in the graph database, build an inverted index between structures and graphs ed r e v Co m a e x b E Step 2. Query Processing to t rm o e T N Mid in the query graph n Enumerate h structures e in t n Calculate the candidate graphs containing these structures n Prune the false positive answers by performing subgraph isomorphism test 39

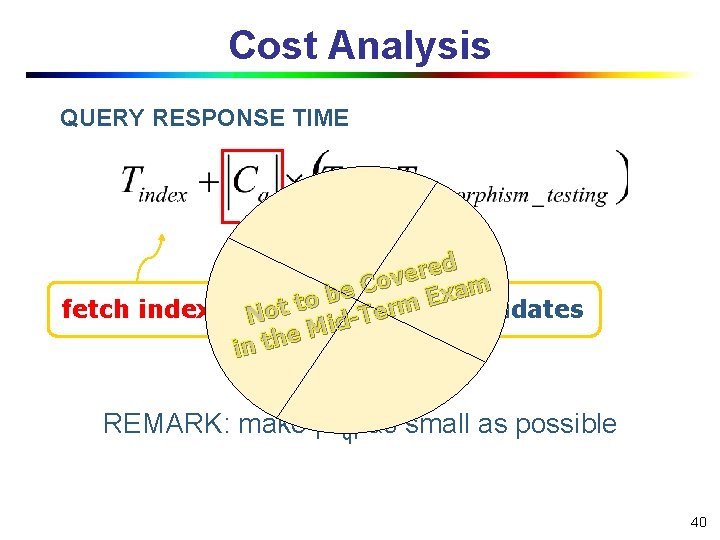
Cost Analysis QUERY RESPONSE TIME ed r e v Co m a e x b E to rm fetch index Not number of candidates e T d Mi e h in t REMARK: make |Cq| as small as possible 40

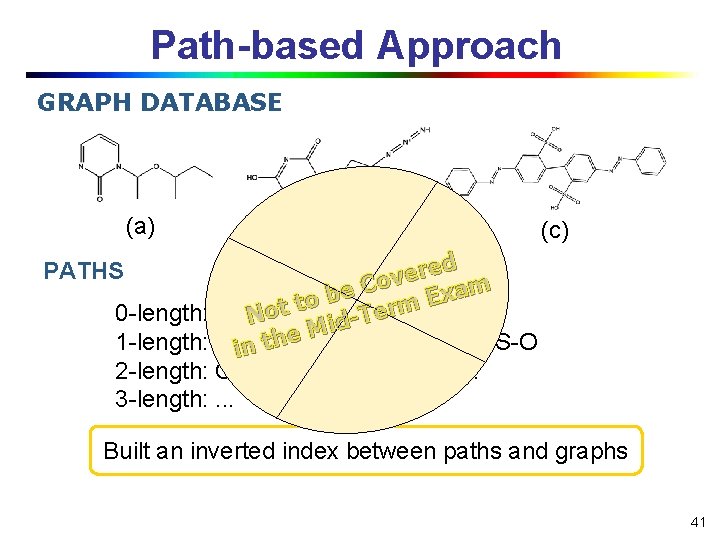
Path-based Approach GRAPH DATABASE (a) (b) (c) ed r e v Co m a e x b E to. S t rm o e 0 -length: C, O, N, T N id M e h 1 -length: C-C, C-N, C-S, N-N, S-O in t. C-O, 2 -length: C-C-C, C-O-C, C-N-C, . . . 3 -length: . . . PATHS Built an inverted index between paths and graphs 41

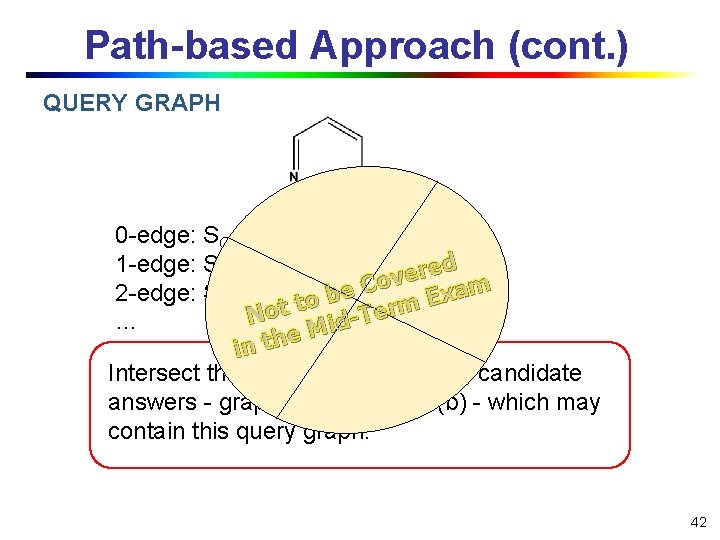
Path-based Approach (cont. ) QUERY GRAPH 0 -edge: SC={a, b, c}, SN={a, b, c} 1 -edge: SC-C={a, b, c}, SC-N={a, db, c} e r e v o C m a e 2 -edge: SC-N-C = {a, b}, … x b E to t rm o e T N … id M e in th Intersect these sets, we obtain the candidate answers - graph (a) and graph (b) - which may contain this query graph. 42

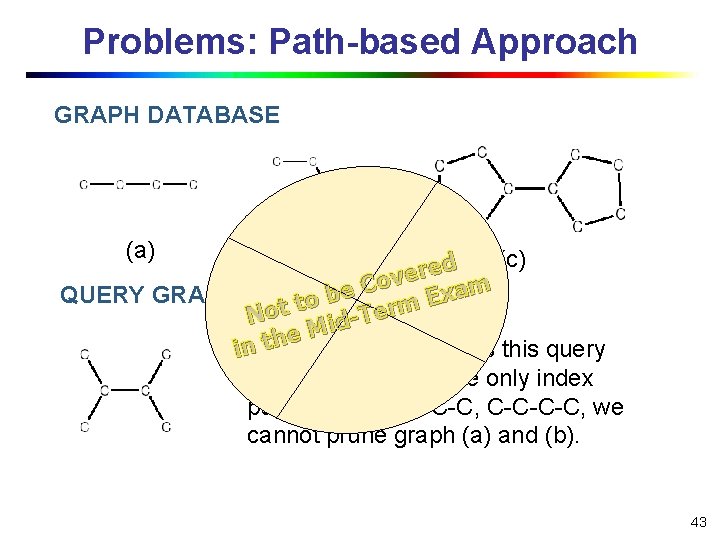
Problems: Path-based Approach GRAPH DATABASE (a) (b) (c) d e r ve o C m a e x QUERY GRAPH b E to t rm o e T N id M e th graph (c) contains this query in. Only graph. However, if we only index paths: C, C-C-C, C-C-C-C, we cannot prune graph (a) and (b). 43

g. Index: Indexing Graphs by Data Mining n Our methodology on graph index: n n n Identify frequent structures in the database, the frequent structures are subgraphs that appear quite often in the graph database ed r e v o Prune redundanttofrequent to maintain xam be C mstructures E t er T No d Mi a small set of discriminative structures e h t n i Create an inverted index between discriminative frequent structures and graphs in the database 44

IDEAS: Indexing with Two Constraints ed r e v Co (~10 x 3 a)m e discriminative b E to m t r o Te N d i e. M h t in frequent (~105) structure (>106) 45

Why Discriminative Subgraphs? Sample database (a) n n (b) (c) ed r e v Co m a e x b E o tstructures: All graphs contain t rm C, C-C-C o e T N id M e Why bother indexing these redundant frequent in th structures? n Only index structures that provide more information than existing structures 46

Discriminative Structures n Pinpoint the most useful frequent structures n Given a set of structures and a new structure , we measure the extra indexing power provided by , ed r e v Co m a e x b E to t rm o e T N id M is small enough, is a discriminative e h t n i n When structure and should be included in the index Index discriminative frequent structures only n Reduce the index size by an order of magnitude 47

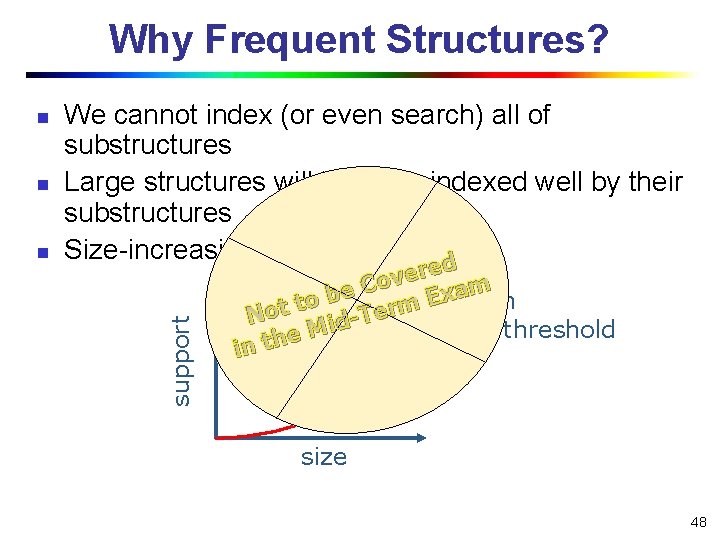
Why Frequent Structures? n n We cannot index (or even search) all of substructures Large structures will likely be indexed well by their substructures Size-increasing support threshold d support n ed r e v Co m a e x b E minimum to m t r o e T N di M support threshold e h t in size 48

Experimental Setting n The AIDS antiviral screen compound dataset from NCI/NIH, containing 43, 905 chemical compounds n Query graphs are randomly extracted from the dataset n ed r e v o C m a e x b Graph. Grep: maximum length (edges) of paths is E to m t r o Te N d i e. M h set at 10 t in n g. Index: maximum size (edges) of structures is set at 10 49

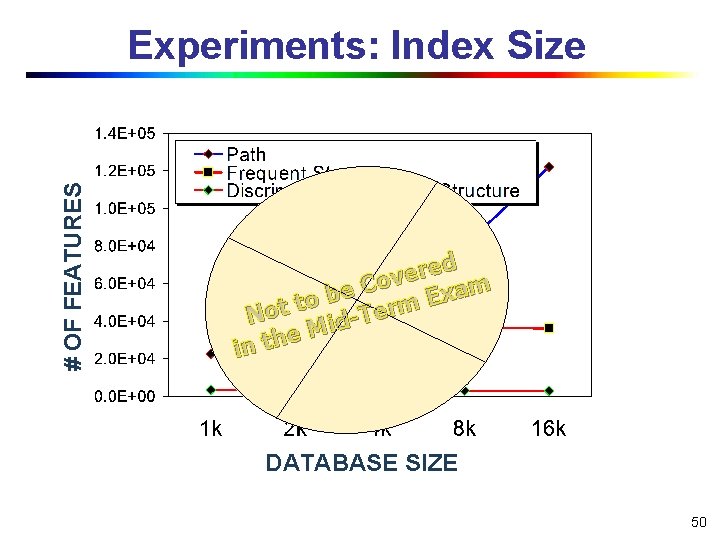
# OF FEATURES Experiments: Index Size ed r e v Co m a e x b E to t rm o e T N id M e in th DATABASE SIZE 50

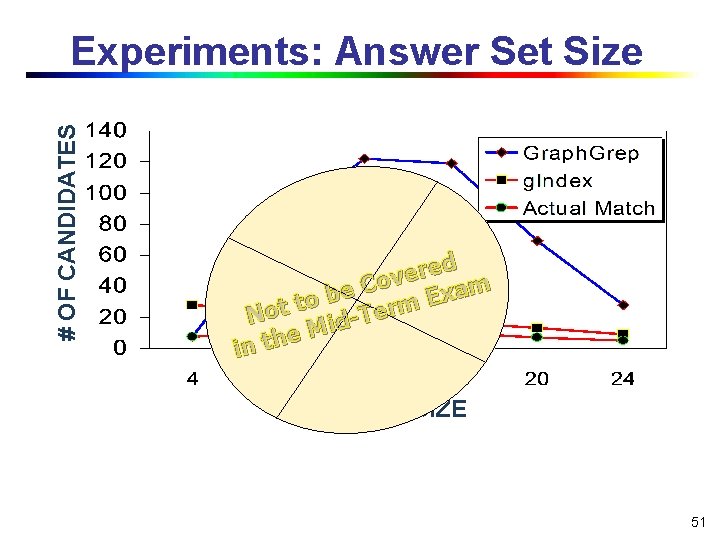
# OF CANDIDATES Experiments: Answer Set Size ed r e v Co m a e x b E to t rm o e T N id M e in th QUERY SIZE 51

Experiments: Incremental Maintenance ed r e v Co m a e x b E to t rm o e T N id M e in th Frequent structures are stable to database updating Index can be built based on a small portion of a graph database, but be used for the whole database

Alternative Graph Indexing Methods n n Graph-structure-based indexing and similarity search n Structure-based index methods, e. g. , g-Index, S-path index n Use index to search for similar graph/network structures Substructure indexing n Key problem: What substructures as indexing features? n g. Index [Yan, Yu & Han, SIGMOD’ 04]: ed Find frequent and r e v Co m mining) discriminative subgraphs (by graph-pattern a e x b E to t rm o e T N n S-path [Zhao & Han, VLDB’ 10]: Use decomposed shortest id M e n th features paths as basic iindexing 53

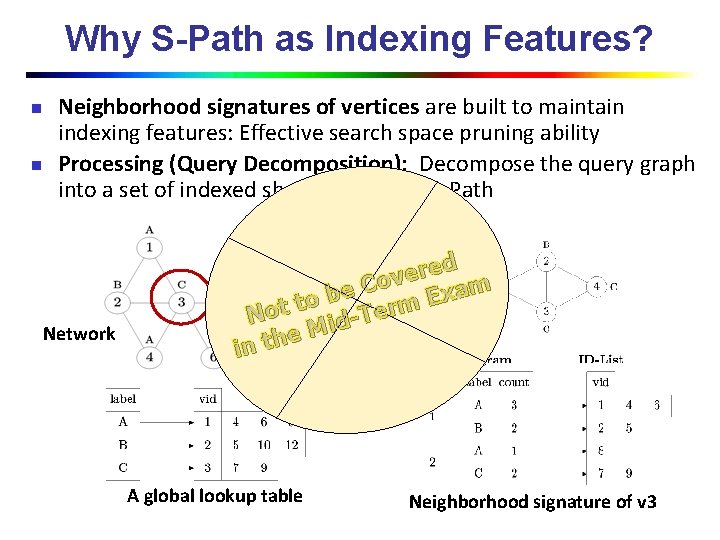
Why S-Path as Indexing Features? n n Neighborhood signatures of vertices are built to maintain indexing features: Effective search space pruning ability Processing (Query Decomposition): Decompose the query graph into a set of indexed shortest paths in S-Path Network ed r e v Co m a e x b E to t rm Query o e T N id M e in th A global lookup table Neighborhood signature of v 3

Graph Mining n Graph Pattern Mining Frequent Subgraph Patterns n Impact on Graph Search I: Graph Indexing n Impact on Graph Search II: d Graph Similarity e r e ov C m a e x b Search E t to rm n er T No d Mi e h Graph Classification (not to be covered) in t n Graph Clustering (not to be covered) n Summary 55

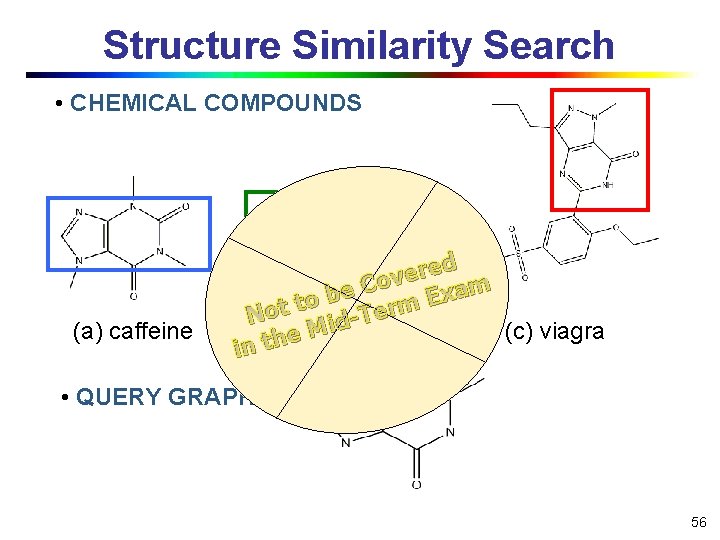
Structure Similarity Search • CHEMICAL COMPOUNDS (a) caffeine ed r e v Co m a e x b E to t rm o e T N id M (b) diurobromine (c) viagra e h t in • QUERY GRAPH 56

Some “Straightforward” Methods n Method 1: Directly compute the similarity between the graphs in the DB and the query graph n Sequential scan Subgraph similarity computation ed r e v Co m from the a Method 2: Form a set of subgraph queries e x b E to t rm o e T N d the exact subgraph search iuse M original query graph and e in th n Costly: If we allow 3 edges to be missed in a 20 -edge n n query graph, it may generate 1, 140 subgraphs 57

Index: Precise vs. Approximate Search n Precise Search n n n Use frequent patterns as indexing features Select features in the database space based on their selectivity n Build the index n Idea: (1) keep the index structure ed r e v Co am e x b E Approximate Searcht to rm e T No d i M e n Hard to build indices covering similar subgraphs— in th explosive number of subgraphs in databases (2) select features in the query space 58

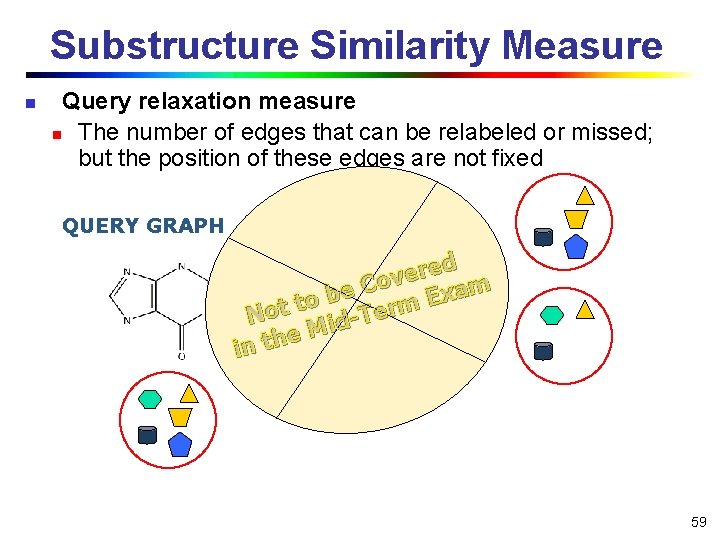
Substructure Similarity Measure n Query relaxation measure n The number of edges that can be relabeled or missed; but the position of these edges are not fixed QUERY GRAPH ed r e v Co m a e x b E to t rm o e T N id M e in th … 59

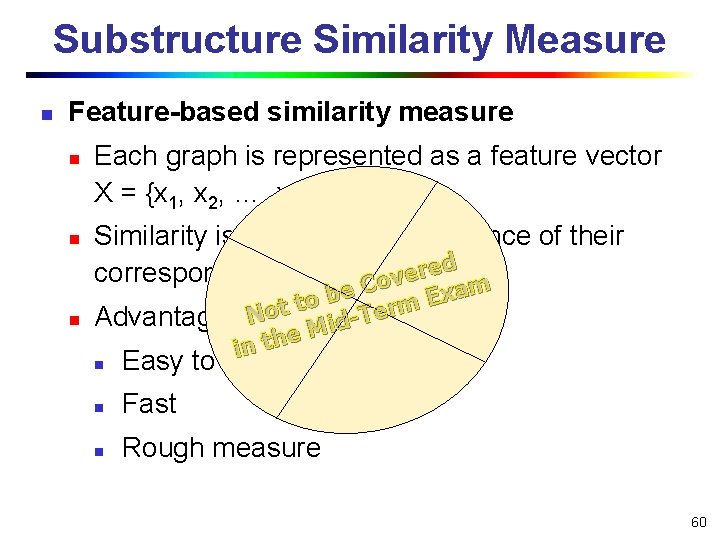
Substructure Similarity Measure n Feature-based similarity measure n n n Each graph is represented as a feature vector X = {x 1, x 2, …, xn} Similarity is defined by the distance of their corresponding vectors. Covered m a x be E o t Advantages Not Mid-Term e h t n i n Easy to index n Fast n Rough measure 60

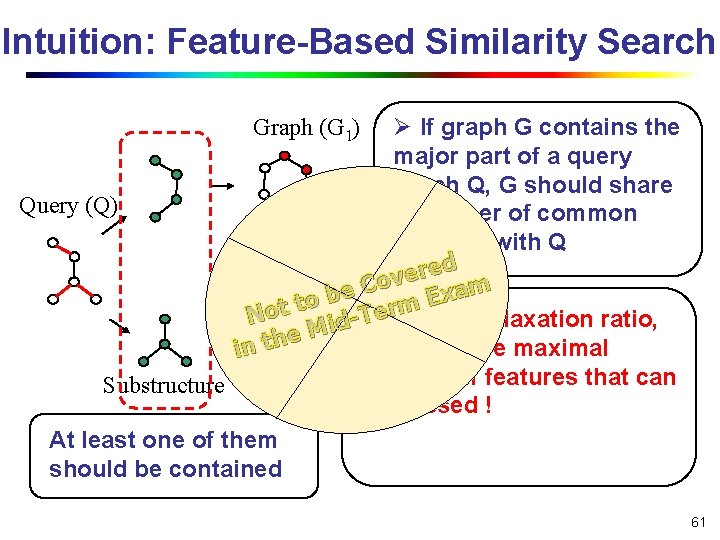
Intuition: Feature-Based Similarity Search Ø If graph G contains the major part of a query graph Q, G should share Query (Q) a number of common features with Q ed r e v Graph (Gb 2 e) Co m a x E to t rm o e T N Ø Given a relaxation ratio, d i M e h t n calculate the maximal i number of features that can Substructure be missed ! At least one of them should be contained Graph (G 1) 61

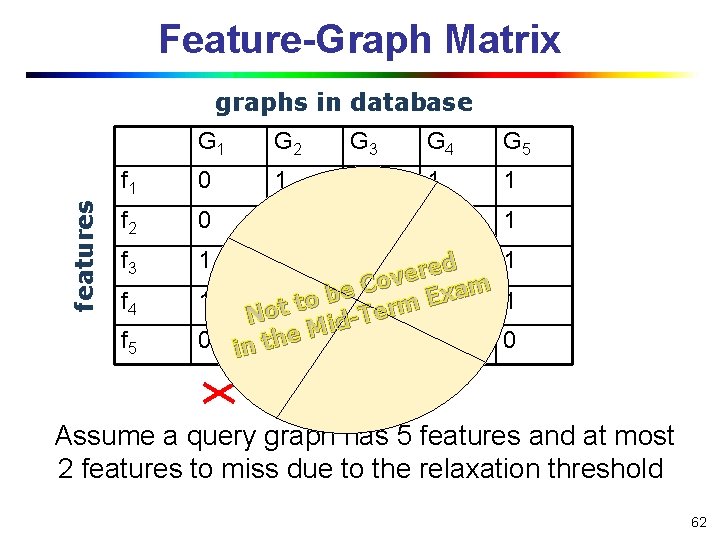
Feature-Graph Matrix features graphs in database G 1 G 2 G 3 G 4 G 5 f 1 0 1 1 f 2 0 1 0 0 1 f 3 1 0 f 4 f 5 1 1 ed 1 r e ov C m a e x b E 1 0 t to 0 erm 0 1 o T N di M 0 in th 0 e 1 1 0 Assume a query graph has 5 features and at most 2 features to miss due to the relaxation threshold 62

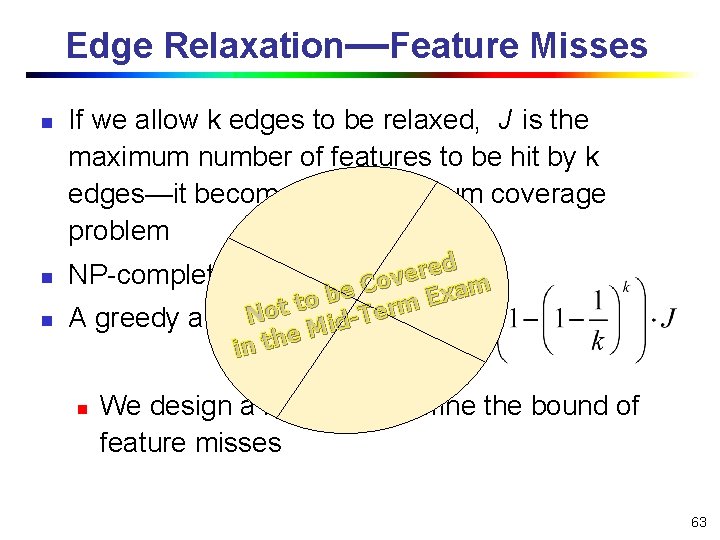
Edge Relaxation—Feature Misses n n n If we allow k edges to be relaxed, J is the maximum number of features to be hit by k edges—it becomes the maximum coverage problem ed r e v Co m a e x b E to t rm o e T N A greedy algorithm exists id M e in th NP-complete n We design a heuristic to refine the bound of feature misses 63

Query Processing Framework n Three steps in processing approximate graph queries Step 1. Index Construction d e r e v Co asxafeatures m n Select small structures in a e b E o t rm ot e T N idand build the featuregraph database, M e th n i graph matrix between the features and the graphs in the database 64

Framework (cont. ) Step 2. Feature Miss Estimation n Determine the indexed features belonging to the query graph ed r e v Cobound m the number n Calculate the upper of a e x b E to t rm o e T N of features that. Mcan id be missed for an e th n i approximate matching, denoted by J n On the query graph, not the graph database 65

Framework (cont. ) Step 3. Query Processing n n Use the feature-graph matrix to edthe number calculate the difference in r e v Co m a e x b E of features. Nobetween G and query t to -Tegraph rm id M e Q, FG – Fi. Qn th If FG – FQ > J, discard G. The remaining graphs constitute a candidate answer set 66

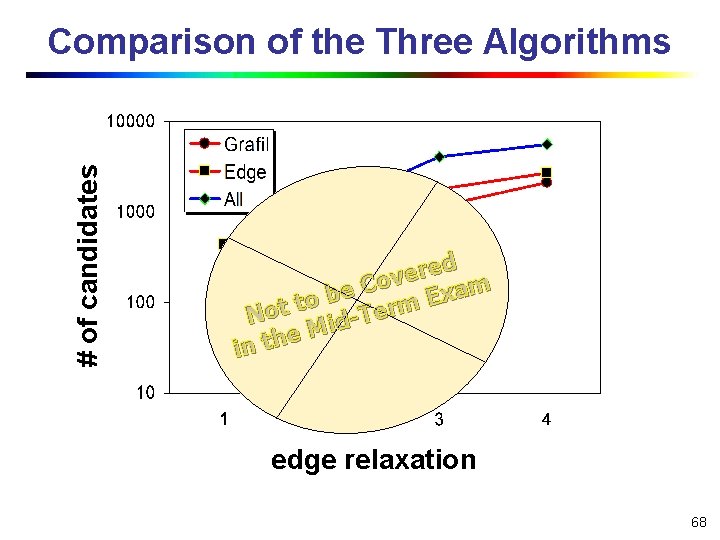
Performance Study n n Database n Chemical compounds of Anti-Aids Drug from NCI/NIH, randomly select 10, 000 compounds Query n Randomly select 30 graphsed with 16 and 20 r e ov C m a e x edges as queryt graphs b E to rm o e T N id M e n Competitive algorithms in th n Grafil: Graph Filter—our algorithm n Edge: use edges only n All: use all the features 67

# of candidates Comparison of the Three Algorithms ed r e v Co m a e x b E to t rm o e T N id M e in th edge relaxation 68

Summary: Graph Pattern Mining n Graph mining has wide applications n Frequent and closed subgraph mining methods n g. Span and Close. Graph: pattern-growth depth-first search approach Graph indexing techniques Covered m a x be E o t rm ot e n Frequent and N discriminative subgraphs are high-quality T d i e. M h t in indexing features n Similarity search in graph databases n n n Indexing and feature-based matching Constraint-based graph pattern mining

References (1) n n n n T. Asai, et al. “Efficient substructure discovery from large semi-structured data”, SDM'02 C. Borgelt and M. R. Berthold, “Mining molecular fragments: Finding relevant substructures of molecules”, ICDM'02 M. Deshpande, M. Kuramochi, and G. Karypis, “Frequent Sub-structure Based Approaches for Classifying Chemical Compounds”, ICDM 2003 M. Deshpande, M. Kuramochi, and G. Karypis. “Automated approaches for classifying structures”, BIOKDD'02 L. Dehaspe, H. Toivonen, and R. King. “Finding frequent substructures in chemical compounds”, KDD'98 C. Faloutsos, K. Mc. Curley, and A. Tomkins, “Fast Discovery of 'Connection Subgraphs”, KDD'04 L. Holder, D. Cook, and S. Djoko. “Substructure discovery in the d subdue system”, KDD'94 e r veand A. Tropsha. J. Huan, W. Wang, D. Bandyopadhyay, J. Snoeyink, C J. o Prins, “Mining spatial motifs from m a e x protein structure graphs”, RECOMB’ 04 o b E t m t o mining er subgraph in the presence of isomorphism”, J. Huan, W. Wang, and J. Prins. N “Efficient of. Tfrequent d Mi ICDM'03 e h t “Mining Coherent Dense Subgraphs across Massive Biological in. J. Zhou, H. Hu, X. Yan, Yu, J. Han and X. Networks for Functional Discovery”, ISMB'05 A. Inokuchi, T. Washio, and H. Motoda. “An apriori-based algorithm for mining frequent substructures from graph data”, PKDD'00 C. James, D. Weininger, and J. Delany. “Daylight Theory Manual Daylight Version 4. 82”. Daylight Chemical Information Systems, Inc. , 2003. G. Jeh, and J. Widom, “Mining the Space of Graph Properties”, KDD'04 M. Koyuturk, A. Grama, and W. Szpankowski. “An efficient algorithm for detecting frequent subgraphs in biological networks”, Bioinformatics, 20: I 200 --I 207, 2004.

References (2) n n n n n M. Kuramochi and G. Karypis. “Frequent subgraph discovery”, ICDM'01 M. Kuramochi and G. Karypis, “GREW: A Scalable Frequent Subgraph Discovery Algorithm”, ICDM’ 04 B. Mc. Kay. Practical graph isomorphism. Congressus Numerantium, 30: 45 --87, 1981. S. Nijssen and J. Kok. A quickstart in frequent structure mining can make a difference. KDD'04 J. Prins, J. Yang, J. Huan, and W. Wang. “Spin: Mining maximal frequent subgraphs from graph databases”. KDD'04 D. Shasha, J. T. -L. Wang, and R. Giugno. “Algorithmics and applications of tree and graph searching”, PODS'02 J. R. Ullmann. “An algorithm for subgraph isomorphism”, J. ACM, 23: 31 --42, 1976. N. Vanetik, E. Gudes, and S. E. Shimony. “Computing frequent graph d patterns from semistructured data”, e r e ICDM'02 ov C m a e x b E C. Wang, W. Wang, J. Pei, Y. Zhu, and B. t. Shi. “Scalable mining of large disk-base graph databases”, KDD'04 o m t r o Te data mining”, SIGKDD Explorations, 5: 59 -68, Nof the art ofid T. Washio and H. Motoda, “State graph-based 2003 e. M h t in X. Yan and J. Han, “g. Span: Graph-Based Substructure Pattern Mining”, ICDM'02 X. Yan and J. Han, “Close. Graph: Mining Closed Frequent Graph Patterns”, KDD'03 X. Yan, P. S. Yu, and J. Han, “Graph Indexing: A Frequent Structure-based Approach”, SIGMOD'04 X. Yan, X. J. Zhou, and J. Han, “Mining Closed Relational Graphs with Connectivity Constraints”, KDD'05 X. Yan, P. S. Yu, and J. Han, “Substructure Similarity Search in Graph Databases”, SIGMOD'05 X. Yan, F. Zhu, J. Han, and P. S. Yu, “Searching Substructures with Superimposed Distance”, ICDE'06 M. J. Zaki. “Efficiently mining frequent trees in a forest”, KDD'02 P. Zhao and J. Han, “On Graph Query Optimization in Large Networks", VLDB'10

05 December 2020 Mining and Searching Graphs in Graph Databases 72