Data mining Data mining Sequence information Mapping information














- Slides: 49

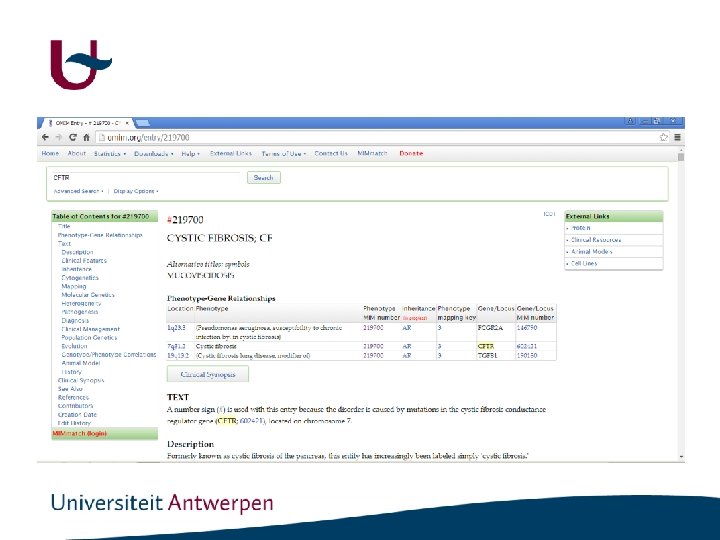
Data mining

Data mining • • • Sequence information Mapping information Phenotypic information Literature Prediction programs - Gene prediction Promotor prediction Functional prediction Structural prediction Variant annotation

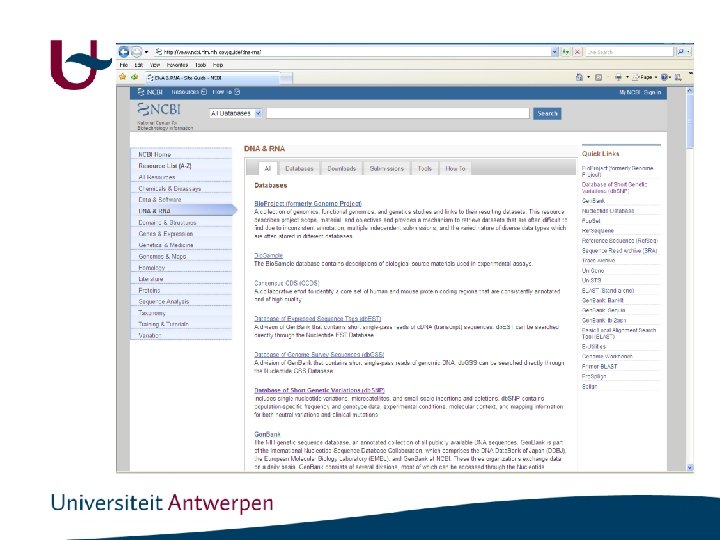
http: //www. ncbi. nlm. nih. gov/








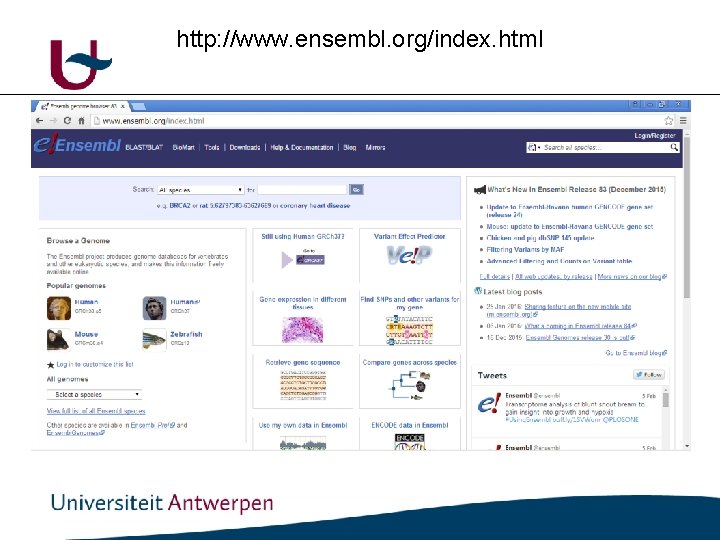
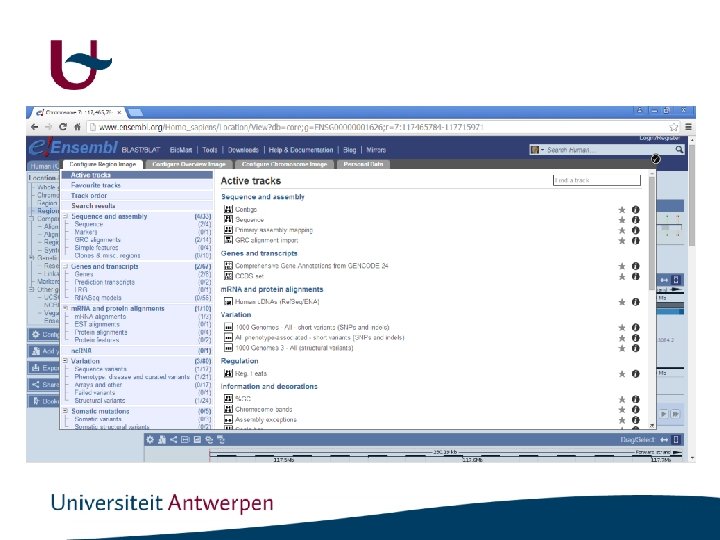
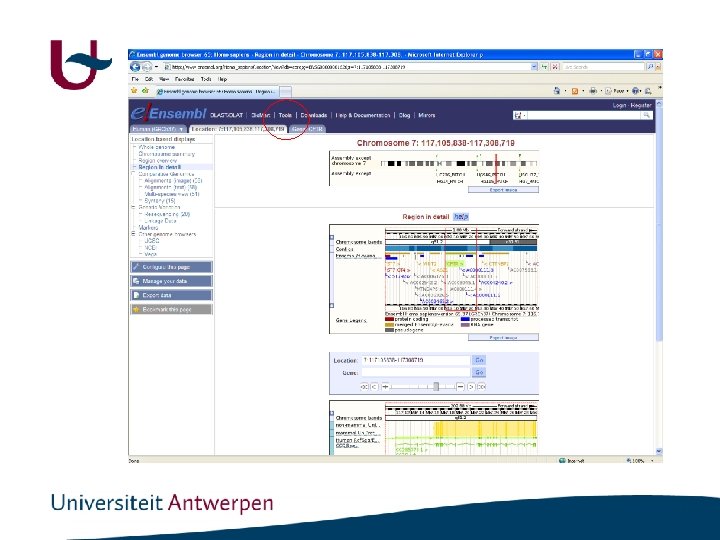
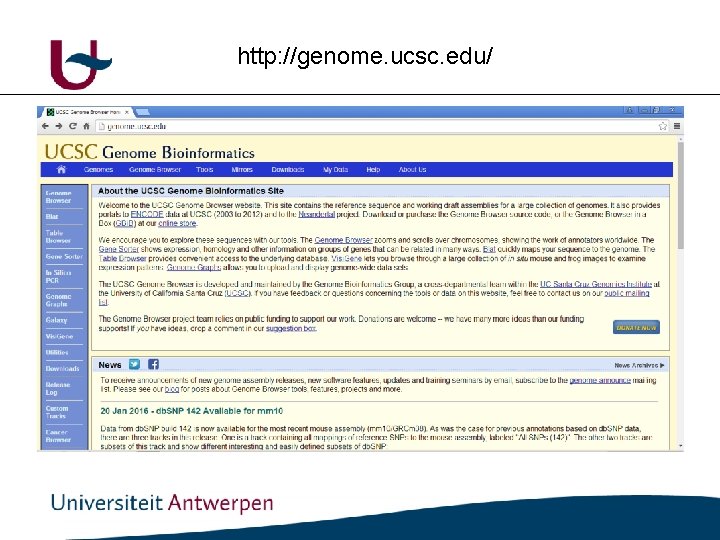
Genome browsers • ENSEMBL: http: //www. ensembl. org/index. html • UCSC : http: //genome. ucsc. edu/

http: //www. ensembl. org/index. html





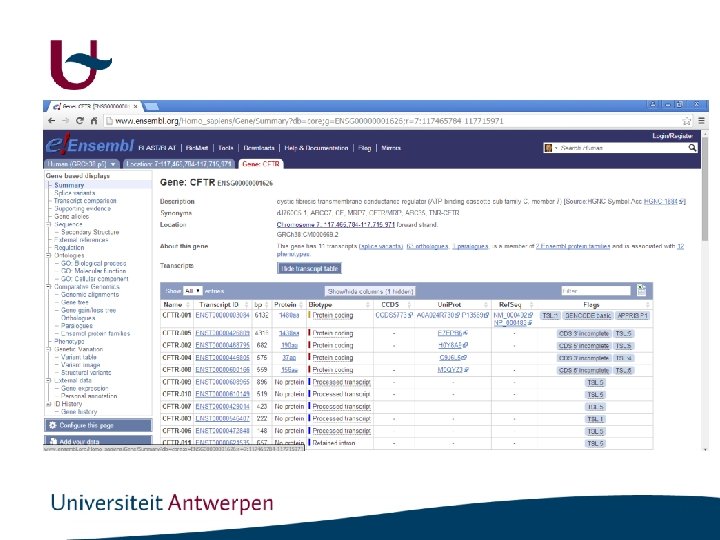
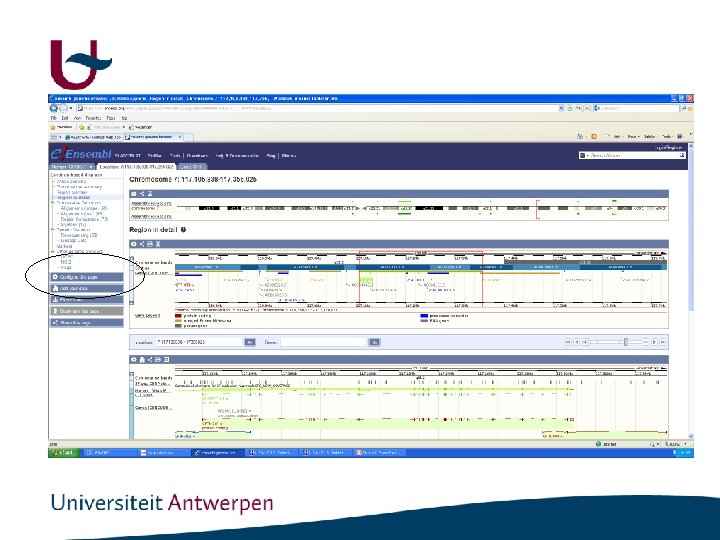
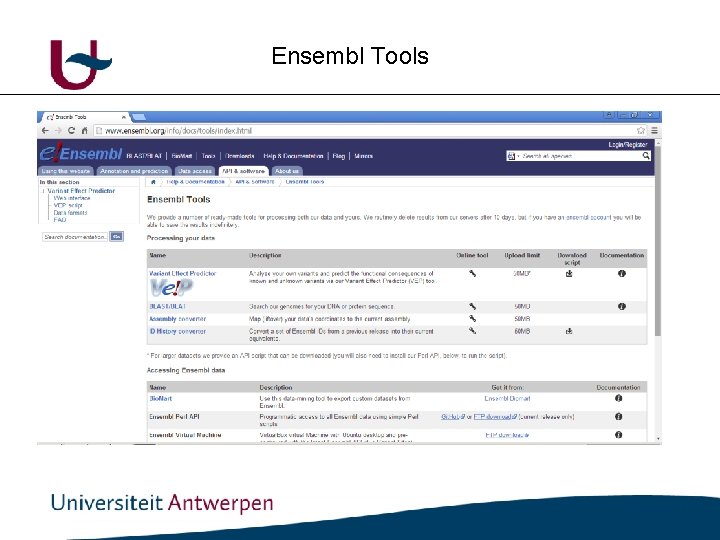
Ensembl Tools

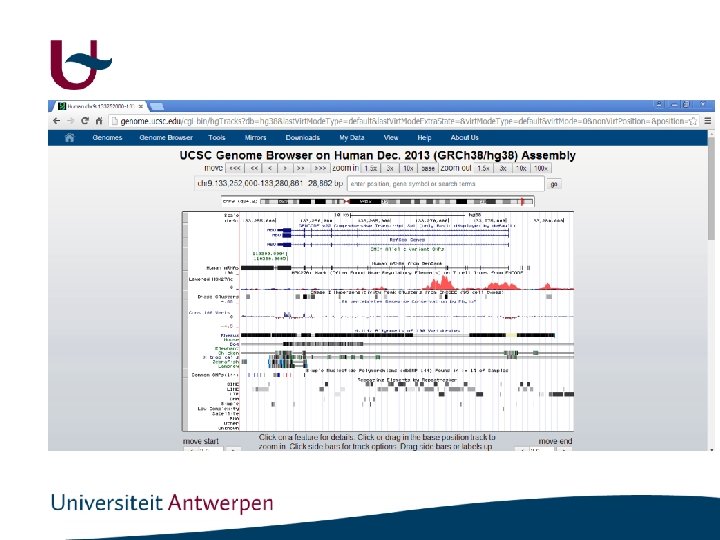
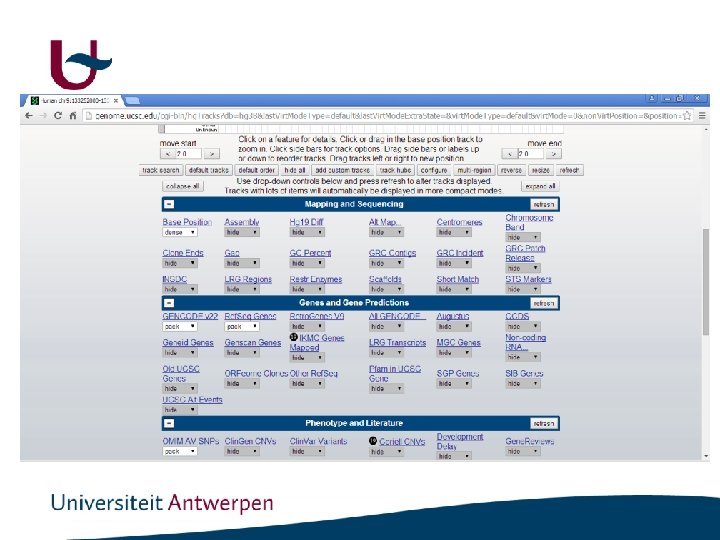
http: //genome. ucsc. edu/



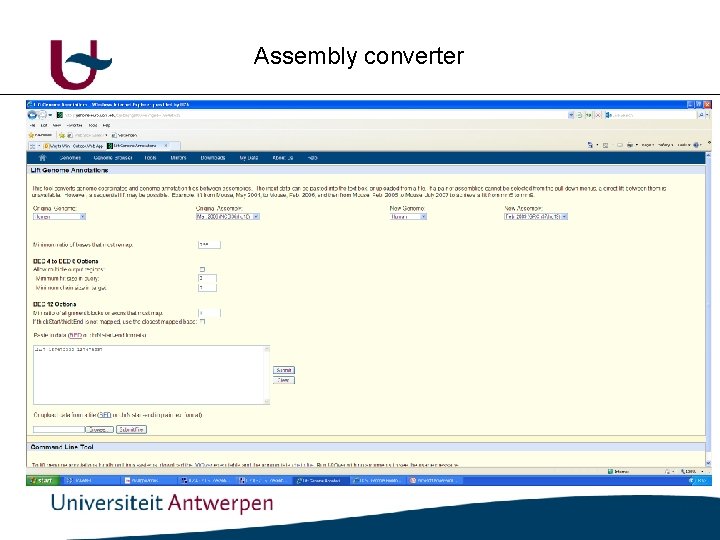
Assembly converter

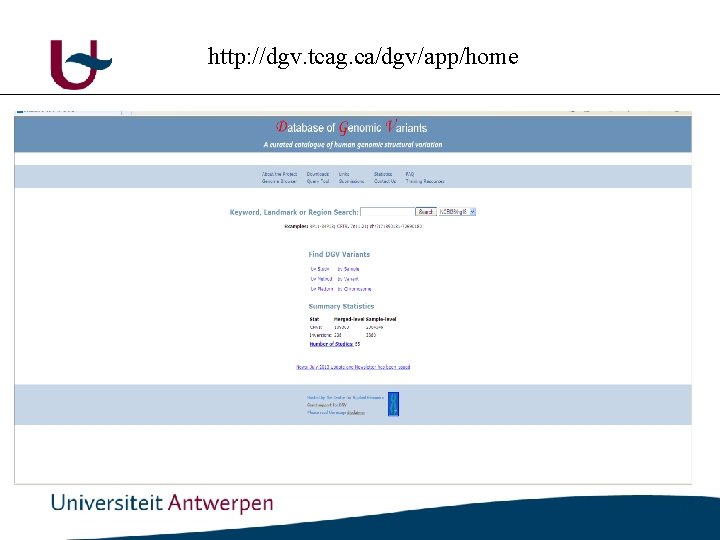
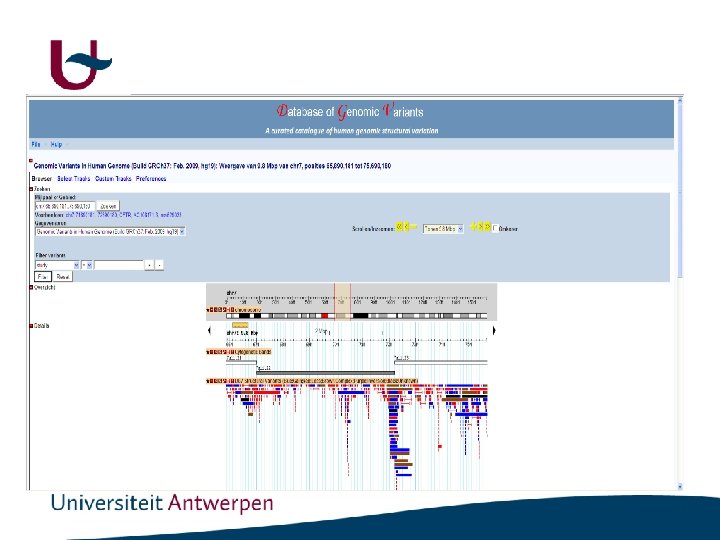
http: //dgv. tcag. ca/dgv/app/home


https: //decipher. sanger. ac. uk/

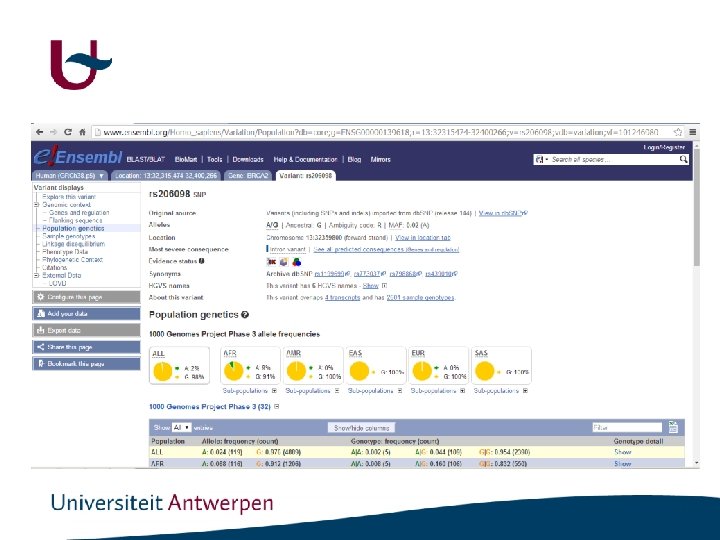
Genomic variant analysis • Genomic variants - Mutation: literature, HGMD, gene specific databases - Polymorphism: db. SNP, EVS, Ex. AC - Prediction programs

http: //www. ncbi. nlm. nih. gov/SNP/



http: //evs. gs. washington. edu/EVS/


Ex. AC (http: //exac. broadinstitute. org/)

Ex. AC

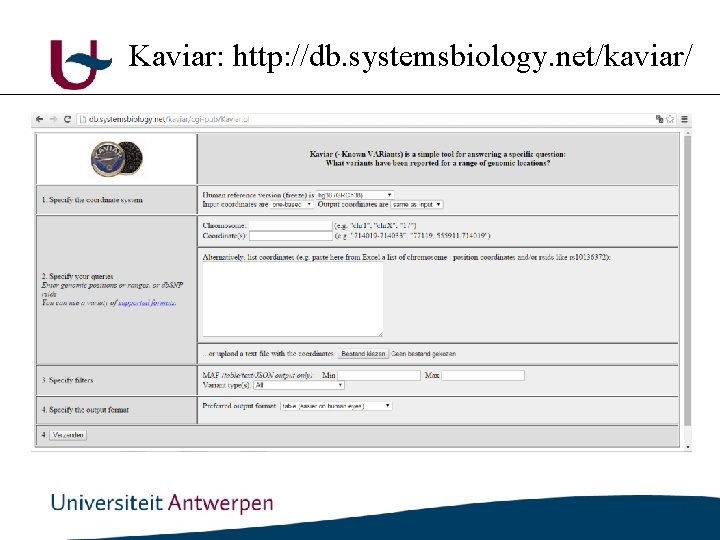
Kaviar: http: //db. systemsbiology. net/kaviar/

Kaviar: http: //db. systemsbiology. net/kaviar/

http: //sift. jcvi. org/

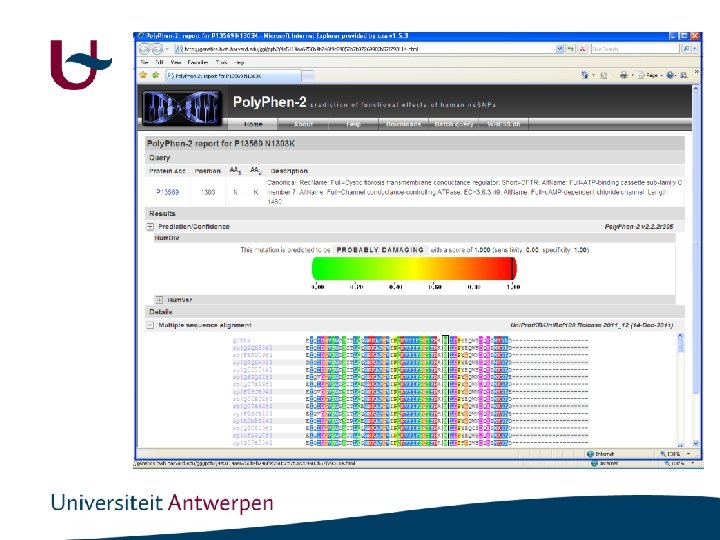
CFTR: p. N 1303 K

CFTR: p. N 1303 K

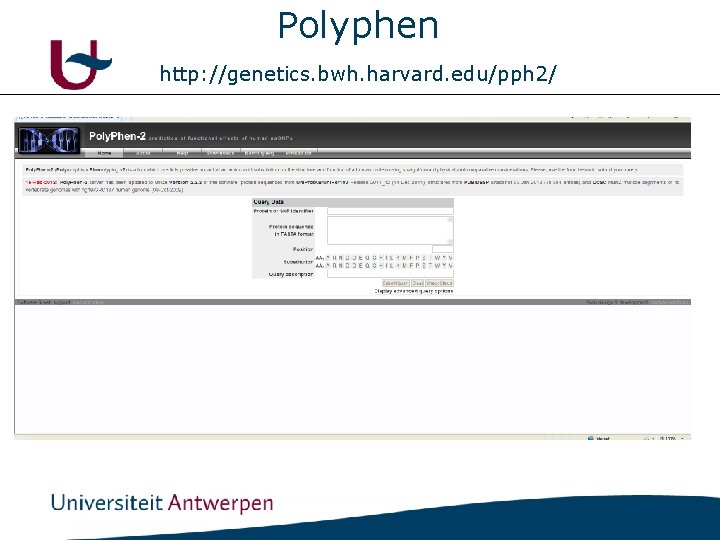
Polyphen http: //genetics. bwh. harvard. edu/pph 2/


Mutation Taster http: //www. mutationtaster. org/

But beware……!!!!! There are many examples of known pathogenic variants predicted to be benign and vice versa

Human Splicing Finder http: //www. umd. be/HSF/

Splice Site Prediction http: //www. fruitfly. org/seq_tools/splice. html

Splice finder programs Gene. Splicer http: //www. cbcb. umd. edu/software /Gene. Splicer/gene_spl. shtml Netgene http: //www. cbs. dtu. dk/services/Net Gene 2/

http: //scholar. google. be

Batch Annotators & Prioritization Gene Panel / Whole Exome Sequencing • Large amounts of variants (50 -50, 000) • Needed information for interpretation: • Location w. r. t. gene (intron/exon/splicing) • Effect on CDS (synonymous/stop-gain/frameshift/. . . ) • Severity of the effect • Function of gene w. r. t. phenotype Example Applications: w. ANNOVAR: http: //wannovar. usc. edu Seattle. Seq: http: //snp. gs. washington. edu/Seattle. Seq. Annotation 141/ VEP: http: //www. ensembl. org/Tools/VEP Variant. DB: http: //biomina. be/apps/variantdb e. Xtasy: http: //extasy. esat. kuleuven. be/ Exomiser: http: //www. sanger. ac. uk/science/tools/exomiser

VEP

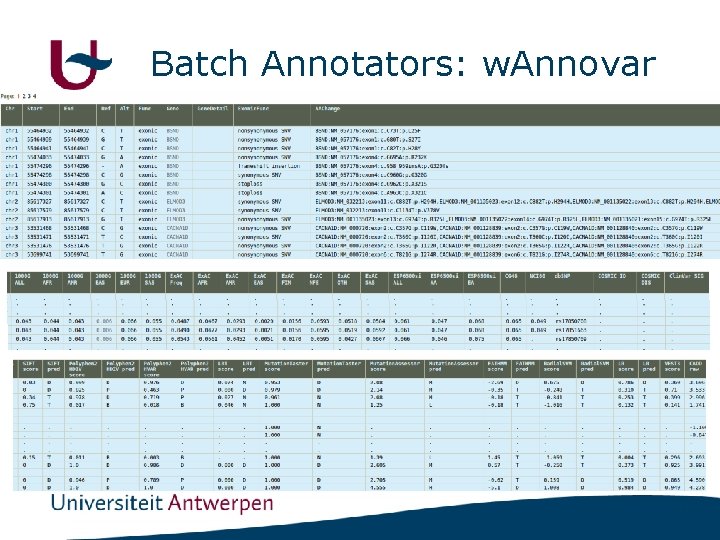
Batch Annotators: w. Annovar

NCBI Ensembl UCSC Genatlas http: //www. ncbi. nlm. nih. gov/ http: //www. ensemble. org/index. html http: //genome. ucsc. edu/ http: //genatlas. medecine. univ-paris 5. fr/ Poly. Phen SIFT Mutation taster Splice prediction http: //genetics. bwh. harvard. edu/pph 2/ http: //sift. jcvi. org/ http: //www. mutationtaster. org/ http: //www. umd. be/HSF/ http: //www. fruitfly. org/seq_tools/splice. html http: //www. cbs. dtu. dk/services/Net. Gene 2/ DECIPHER https: //decipher. sanger. ac. uk/