DAL for theorists Implementation of the SNAP service

- Slides: 12

DAL for theorists: Implementation of the SNAP service for the TVO Claudio Gheller, Giuseppe Fiameni Inter. Universitary Computing Center CINECA, Bologna Ugo Becciani, Alessandro Costa Astrophysical Observatory of Catania Victoria, May 2006

The Simple Numerical Access Protocol Service The Snap service extracts or "cuts out" rectangular (spherical or even irregular) regions of some larger theory dataset, returning a subset of the requested size to the client. Snap basic components: • DATA • SNAP code • SERVICE Victoria, May 2006

1. Data and Data Model In order to analyze the needs of data produced by numerical simulations, we have considered a wide spectrum of applications: • Particle based Cosmological simulations • Grid based Cosmological simulations • Magnatohydrodynamics simulations • Planck mission simulated data • . . . (thanks to V. Antonuccio, G. Bodo, S. Borgani, N. Lanza, L. Tornatore) At the moment, we consider only RAW data Victoria, May 2006

1. Data In general, data produced by numerical simulations are • Large (GB to TB scale) • Monolithic (few files contains plenty of data) • Uncompressible • Non standard (propretary formats are the rule) • Non portable (depend from simulation machine) • No (or few) annotations – metadata • Heterogeneous in units (often code units) Victoria, May 2006

Data: the HDF 5 format HDF 5 (http: //hdf. ncsa. uiuc. edu) represents a possible solution to deal with such data HDF 5 is • Portable between most of modern platform • High performance • Well supported • Well documented • Rich of tools HDF 5 data files are • Platform independent (portable) • Well organized • Self defined • Metadata enriched • Efficiently accessible HDF 5 drawbacks • Requires some expertise and skill to be used • Information are difficult to access • Can be subject to major library changes (see HDF 4 to HDF 5) Victoria, May 2006

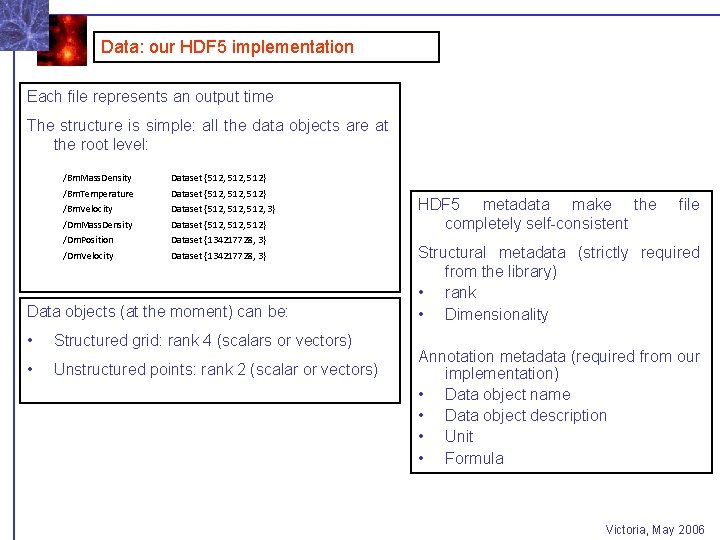
Data: our HDF 5 implementation Each file represents an output time The structure is simple: all the data objects are at the root level: /Bm. Mass. Density Dataset {512, 512} /Bm. Temperature Dataset {512, 512} /Bm. Velocity Dataset {512, 3} /Dm. Mass. Density Dataset {512, 512} /Dm. Position Dataset {134217728, 3} /Dm. Velocity Dataset {134217728, 3} Data objects (at the moment) can be: • Structured grid: rank 4 (scalars or vectors) • Unstructured points: rank 2 (scalar or vectors) HDF 5 metadata make the completely self-consistent file Structural metadata (strictly required from the library) • rank • Dimensionality Annotation metadata (required from our implementation) • Data object name • Data object description • Unit • Formula Victoria, May 2006

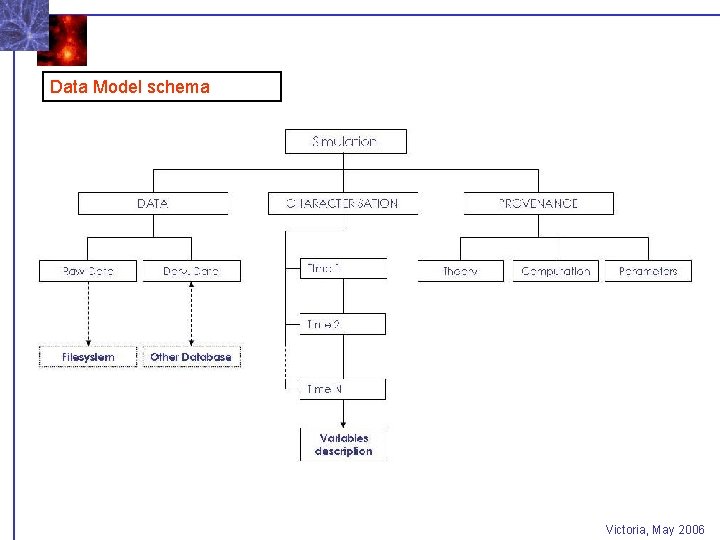
Data Model schema Victoria, May 2006

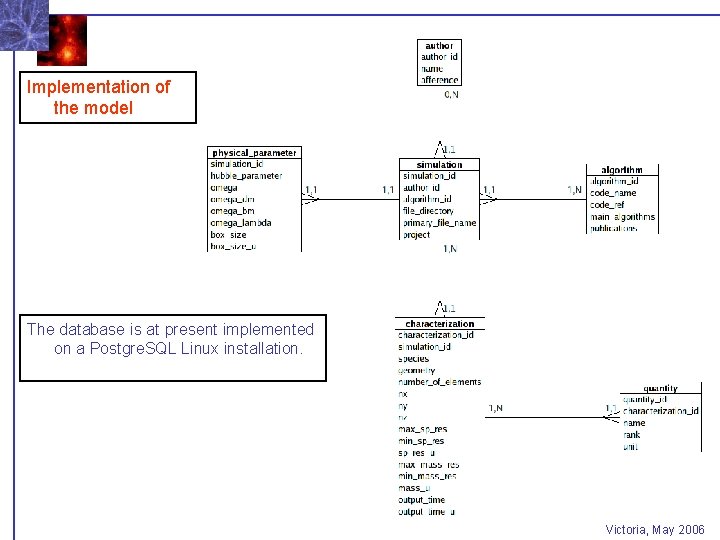
Implementation of the model The database is at present implemented on a Postgre. SQL Linux installation. Victoria, May 2006

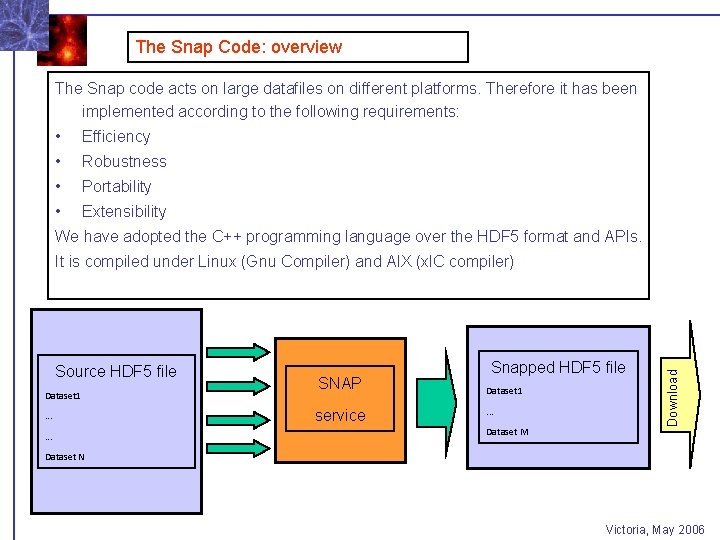
The Snap Code: overview The Snap code acts on large datafiles on different platforms. Therefore it has been implemented according to the following requirements: • Efficiency • Robustness • Portability • Extensibility We have adopted the C++ programming language over the HDF 5 format and APIs. Source HDF 5 file Dataset 1. . . SNAP service Snapped HDF 5 file Dataset 1. . . Dataset M Download It is compiled under Linux (Gnu Compiler) and AIX (xl. C compiler) Dataset N Victoria, May 2006

The Snap Code Goal: select all the data that fall inside a pre-defined region. At present the region can be only rectangular. Input: Output: Data filename One ore more HDF 5 file with the same descriptive metadata as the original dataset. Data objects (one or more) Spatial Units Box Center Box Size Output filename Data objects names Victoria, May 2006

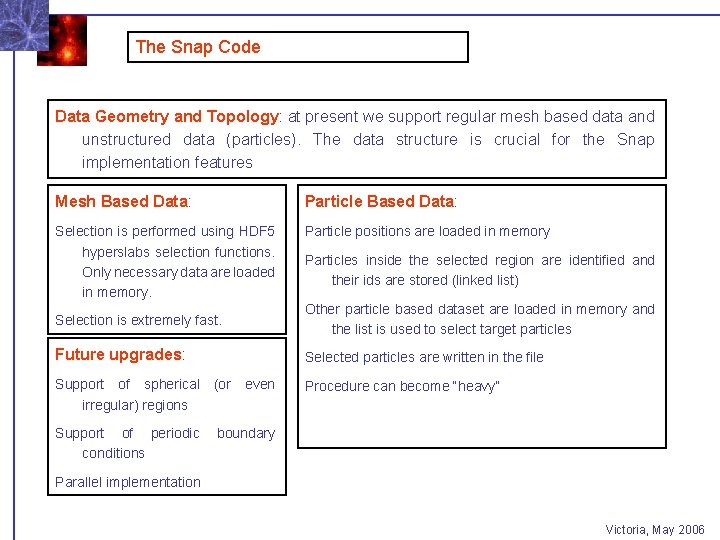
The Snap Code Data Geometry and Topology: at present we support regular mesh based data and unstructured data (particles). The data structure is crucial for the Snap implementation features Mesh Based Data: Particle Based Data: Selection is performed using HDF 5 hyperslabs selection functions. Only necessary data are loaded in memory. Particle positions are loaded in memory Particles inside the selected region are identified and their ids are stored (linked list) Selection is extremely fast. Other particle based dataset are loaded in memory and the list is used to select target particles Future upgrades: Selected particles are written in the file Support of spherical (or even irregular) regions Procedure can become “heavy” Support of periodic conditions boundary Parallel implementation Victoria, May 2006

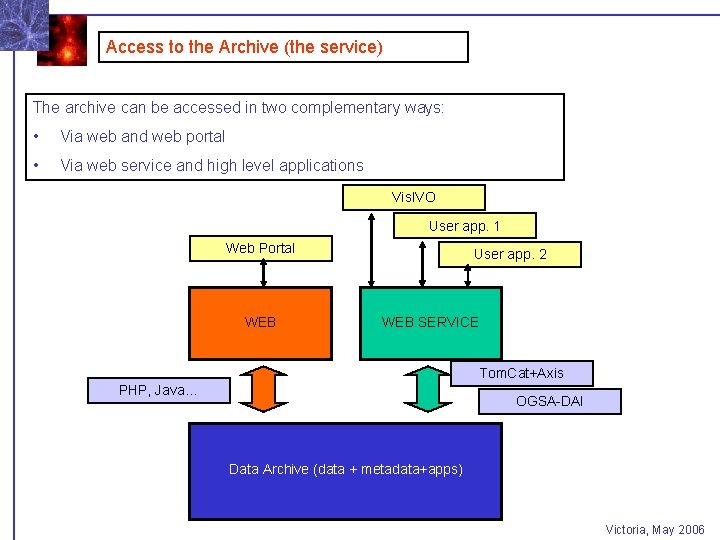
Access to the Archive (the service) The archive can be accessed in two complementary ways: • Via web and web portal • Via web service and high level applications Vis. IVO User app. 1 Web Portal WEB User app. 2 WEB SERVICE Tom. Cat+Axis PHP, Java… OGSA-DAI Data Archive (data + metadata+apps) Victoria, May 2006