Current status of genomic evaluation for U S









- Slides: 47

Current status of genomic evaluation for U. S. dairy cattle Dr. George R. Wiggans Animal Improvement Programs Laboratory Agricultural Research Service, USDA Beltsville, MD 20705 -2350 301 -504 -8407 (voice) 301 -504 -8092 (fax) george. wiggans@ars. usda. gov China Emerging Markets Program Seminar Wiggans, 2013

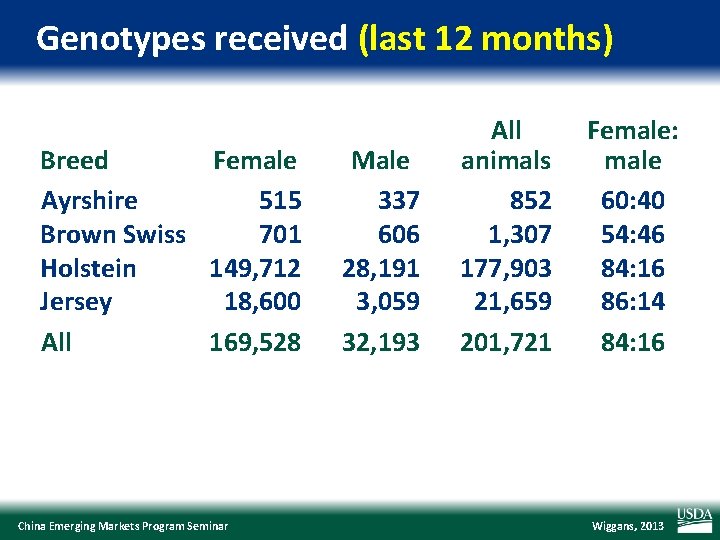
Genotypes received (last 12 months) Breed Female Ayrshire 515 Brown Swiss 701 Holstein 149, 712 Jersey 18, 600 All 169, 528 China Emerging Markets Program Seminar Male 337 606 28, 191 3, 059 32, 193 All animals 852 1, 307 177, 903 21, 659 201, 721 Female: male 60: 40 54: 46 84: 16 86: 14 84: 16 Wiggans, 2013

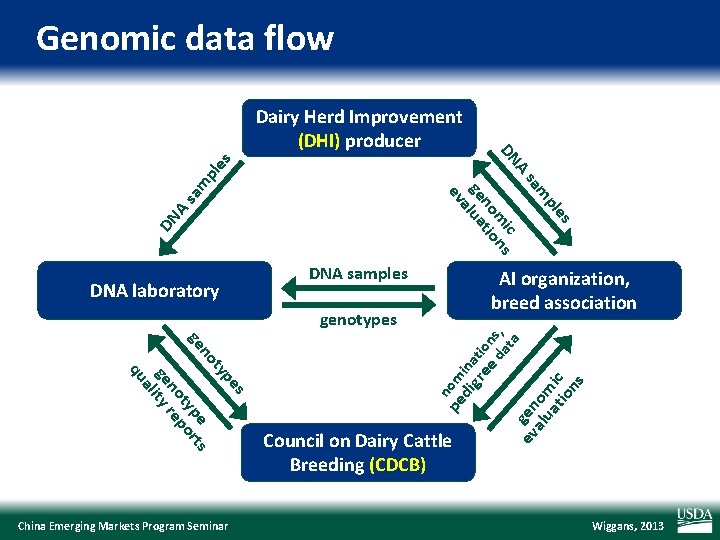
Genomic data flow DN A sa m pl es es pl m sa A ic DN m ns no tio ge lua a ev Dairy Herd Improvement (DHI) producer AI organization, breed association a at s pe ty no pe rts ge ty po no re ge lity a qu China Emerging Markets Program Seminar pe nom di ina gr tio ee n d s, genotypes Council on Dairy Cattle Breeding (CDCB) ev gen alu om at ic io ns DNA laboratory DNA samples Wiggans, 2013

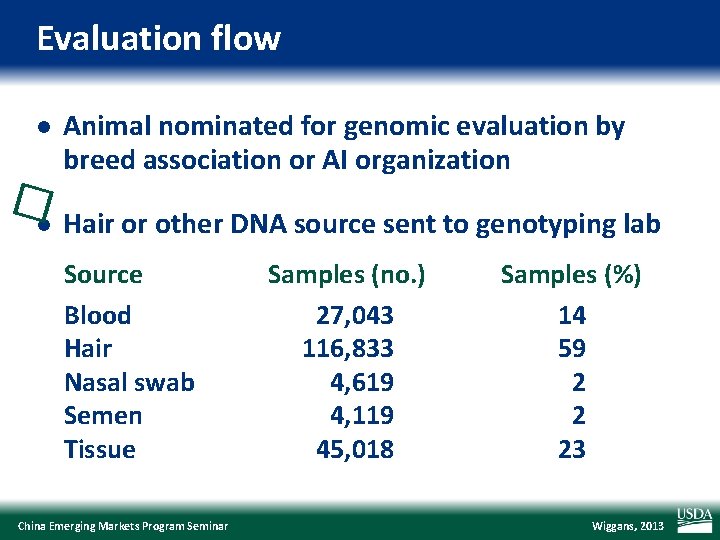
Evaluation flow l Animal nominated for genomic evaluation by breed association or AI organization �l Hair or other DNA source sent to genotyping lab Source Blood Hair Nasal swab Semen Tissue China Emerging Markets Program Seminar Samples (no. ) 27, 043 116, 833 4, 619 4, 119 45, 018 Samples (%) 14 59 2 2 23 Wiggans, 2013

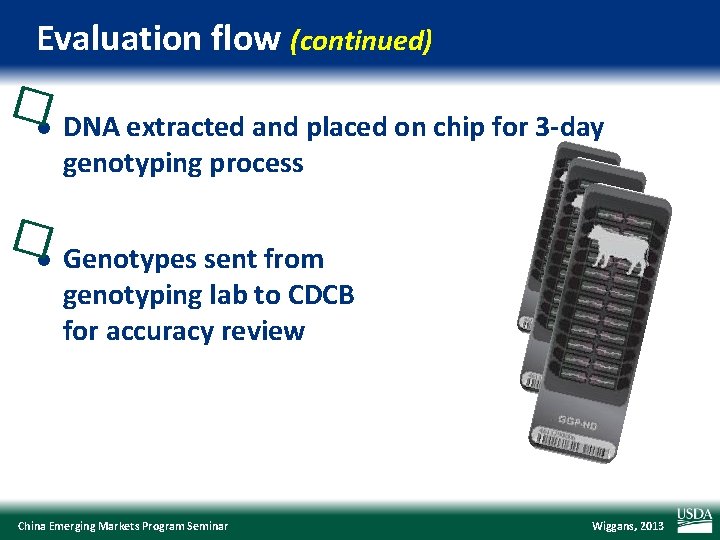
Evaluation flow (continued) �l DNA extracted and placed on chip for 3 -day genotyping process �l Genotypes sent from genotyping lab to CDCB for accuracy review China Emerging Markets Program Seminar Wiggans, 2013

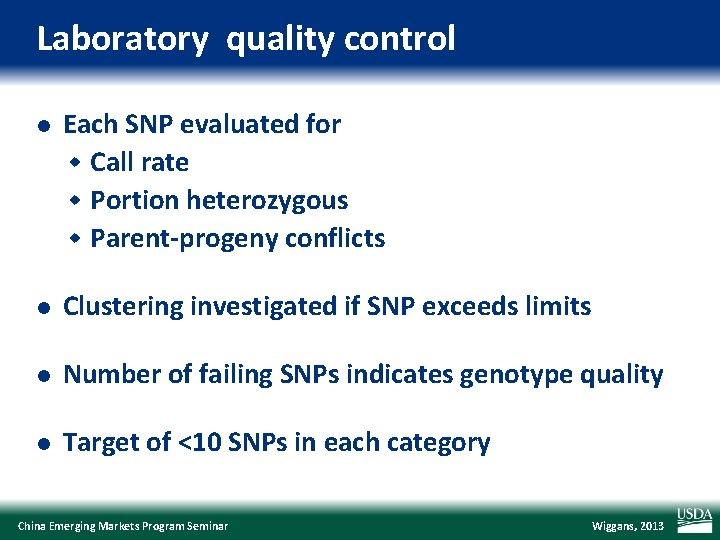
Laboratory quality control l Each SNP evaluated for w Call rate w Portion heterozygous w Parent-progeny conflicts l Clustering investigated if SNP exceeds limits l Number of failing SNPs indicates genotype quality l Target of <10 SNPs in each category China Emerging Markets Program Seminar Wiggans, 2013

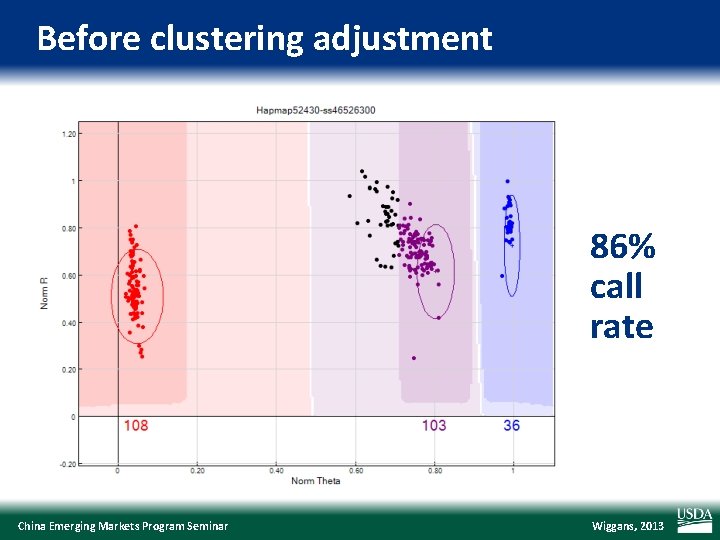
Before clustering adjustment 86% call rate China Emerging Markets Program Seminar Wiggans, 2013

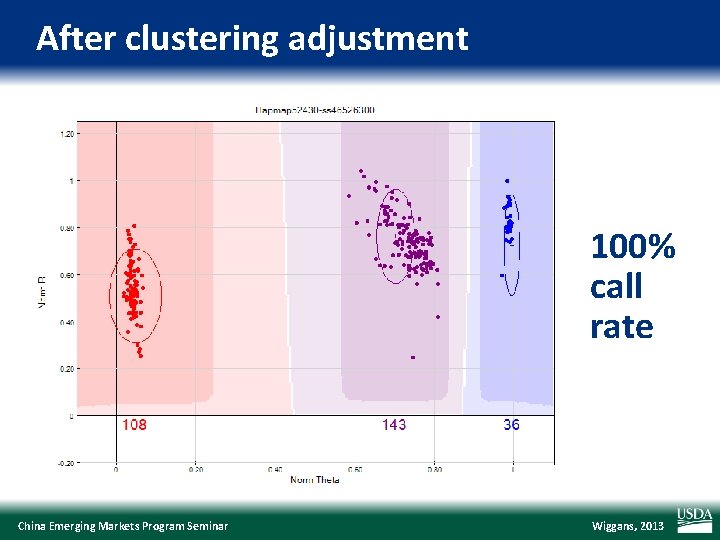
After clustering adjustment 100% call rate China Emerging Markets Program Seminar Wiggans, 2013

Evaluation flow (continued) � l � l � l Genotype calls modified as necessary Genotypes loaded into database Nominators receive reports of parentage and other conflicts Pedigree or animal assignments corrected Genotypes extracted and imputed to 45 K China Emerging Markets Program Seminar Wiggans, 2013

Imputation l Based on splitting genotype into individual chromosomes (maternal and paternal contributions) l Missing SNPs assigned by tracking inheritance from ancestors and descendants l Imputed dams increase predictor population l Genotypes from all chips merged by imputing SNPs not present China Emerging Markets Program Seminar Wiggans, 2013

findhap l Developed by Dr. Paul Van. Raden, ARS, USDA l Divides chromosomes into segments l Allows for successively shorter segments (usually 3 runs) w Long segments lock in identical by descent w Shorter segments fill in missing SNPs l Separates genotype into maternal and paternal contribution, haplotypes (phasing) l Builds haplotype library sequenced by frequency China Emerging Markets Program Seminar Wiggans, 2013

Evaluation flow (continued) � l SNP effects estimated � l Final evaluations calculated � l Evaluations released to dairy industry w Download from CDCB FTP site with separate files for each nominator w Monthly release for new animals w All genomic evaluations updated 3 times each year with traditional evaluations China Emerging Markets Program Seminar Wiggans, 2013

Information sources for evaluations l Traditional evaluations of genotyped bulls and cows used to estimate SNP effects l Combined final evaluation w Sum of SNP effects for an animal’s alleles w Polygenetic effect w Traditional evaluation l Pedigree data used and validated by genotypes China Emerging Markets Program Seminar Wiggans, 2013

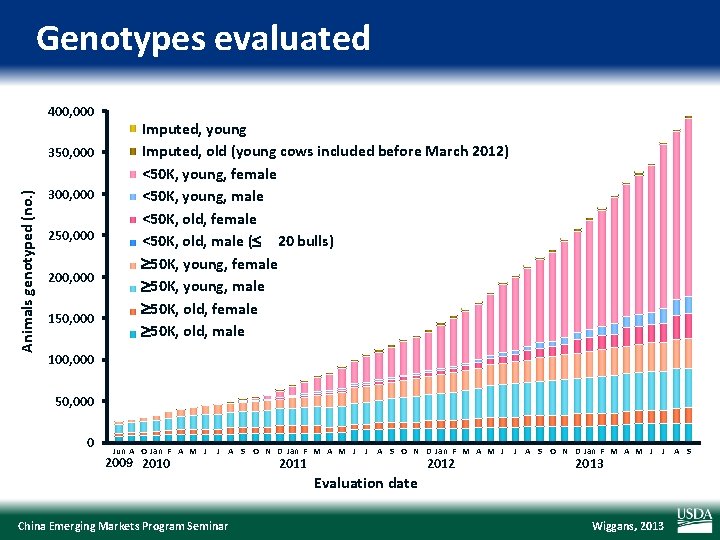
Genotypes evaluated 400, 000 350, 000 Young imputed Imputed, young Old imputed Imputed, old (young cows included before March 2012) Animals genotyped (no. ) <50 K, young, female Female Young <50 K 300, 000 250, 000 200, 000 150, 000 <50 K, Young young, male Male <50 K, old, female Female Old <50 K, Old old, male ( Male <50 K 20 bulls) 50 K, young, female Female Young >=50 K 50 K, Young young, male Male >=50 K 50 K, old, female Female Old >=50 K 50 K, Old old, male Male >=50 K 100, 000 50, 000 0 Jun A O Jan F A M J 2009 2010 J A S O N D Jan F M A M J 2011 J A S O N D Jan F M A M J 2012 J A S O N D Jan F M A M J 2013 J A S Evaluation date China Emerging Markets Program Seminar Wiggans, 2013

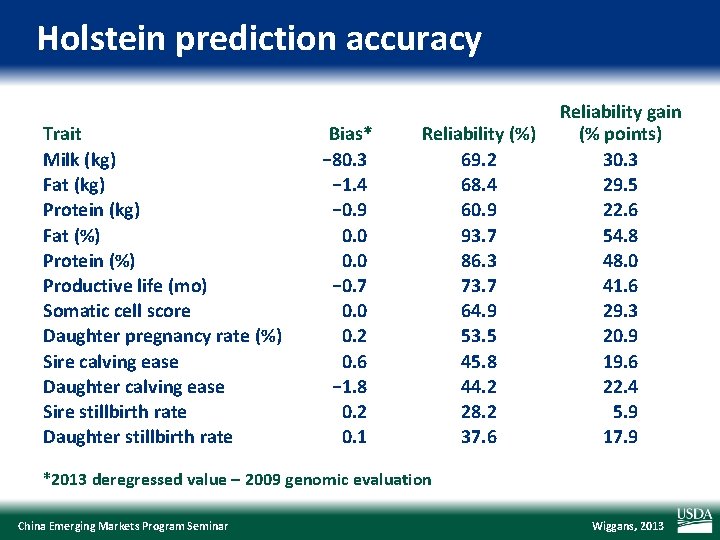
Holstein prediction accuracy Trait Milk (kg) Fat (kg) Protein (kg) Fat (%) Protein (%) Productive life (mo) Somatic cell score Daughter pregnancy rate (%) Sire calving ease Daughter calving ease Sire stillbirth rate Daughter stillbirth rate Bias* − 80. 3 − 1. 4 − 0. 9 0. 0 − 0. 7 0. 0 0. 2 0. 6 − 1. 8 0. 2 0. 1 Reliability (%) 69. 2 68. 4 60. 9 93. 7 86. 3 73. 7 64. 9 53. 5 45. 8 44. 2 28. 2 37. 6 Reliability gain (% points) 30. 3 29. 5 22. 6 54. 8 48. 0 41. 6 29. 3 20. 9 19. 6 22. 4 5. 9 17. 9 *2013 deregressed value – 2009 genomic evaluation China Emerging Markets Program Seminar Wiggans, 2013

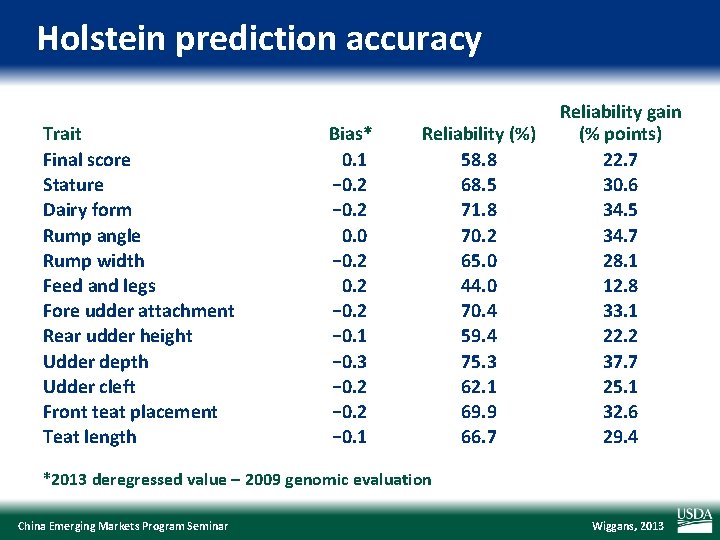
Holstein prediction accuracy Trait Final score Stature Dairy form Rump angle Rump width Feed and legs Fore udder attachment Rear udder height Udder depth Udder cleft Front teat placement Teat length Bias* 0. 1 − 0. 2 0. 0 − 0. 2 − 0. 1 − 0. 3 − 0. 2 − 0. 1 Reliability (%) 58. 8 68. 5 71. 8 70. 2 65. 0 44. 0 70. 4 59. 4 75. 3 62. 1 69. 9 66. 7 Reliability gain (% points) 22. 7 30. 6 34. 5 34. 7 28. 1 12. 8 33. 1 22. 2 37. 7 25. 1 32. 6 29. 4 *2013 deregressed value – 2009 genomic evaluation China Emerging Markets Program Seminar Wiggans, 2013

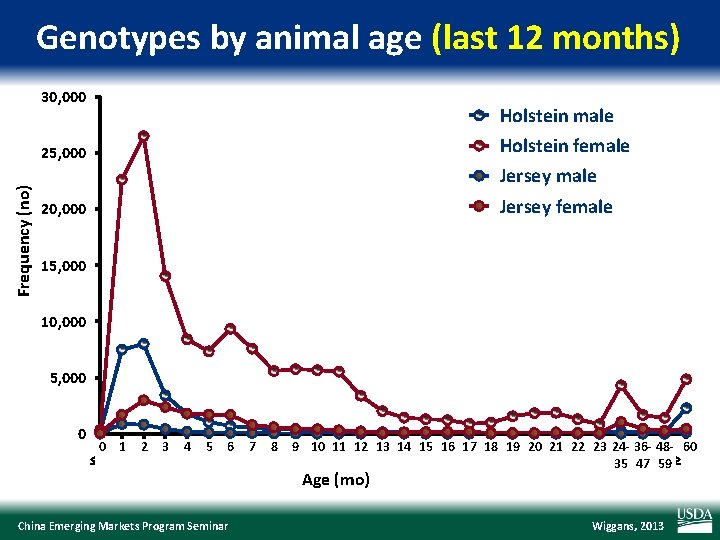
Genotypes by animal age (last 12 months) 30, 000 Holstein male Holstein female Frequency (no) 25, 000 Jersey male Jersey female 20, 000 15, 000 10, 000 5, 000 0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 - 36 - 48 - 60 35 47 59 Age (mo) China Emerging Markets Program Seminar Wiggans, 2013

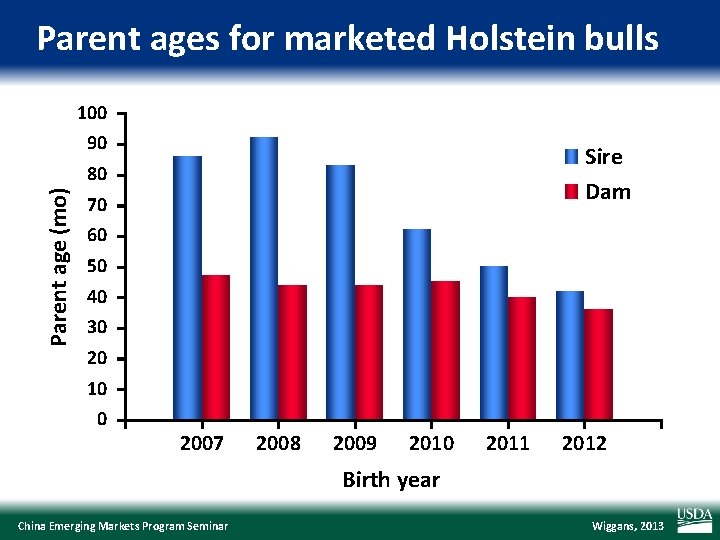
Parent ages for marketed Holstein bulls 100 90 Sire Dam Parent age (mo) 80 70 60 50 40 30 20 10 0 2007 2008 2009 2010 2011 2012 Birth year China Emerging Markets Program Seminar Wiggans, 2013

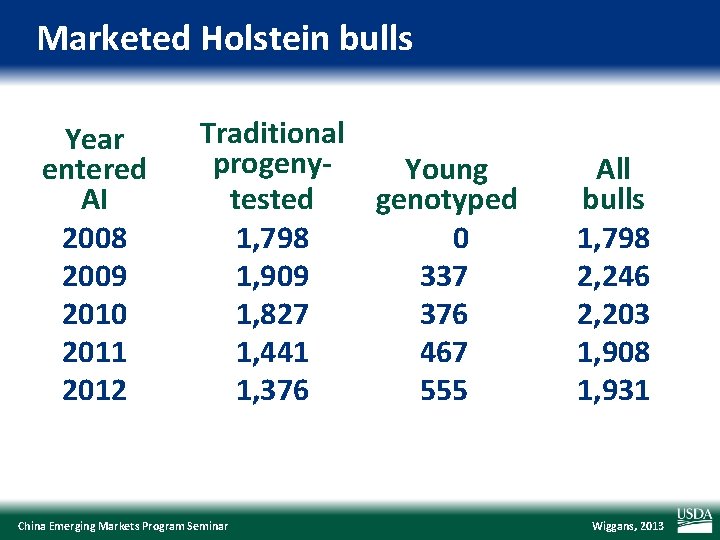
Marketed Holstein bulls Year entered AI 2008 2009 2010 2011 2012 Traditional progeny. Young tested genotyped 1, 798 0 1, 909 337 1, 827 376 1, 441 467 1, 376 555 China Emerging Markets Program Seminar All bulls 1, 798 2, 246 2, 203 1, 908 1, 931 Wiggans, 2013

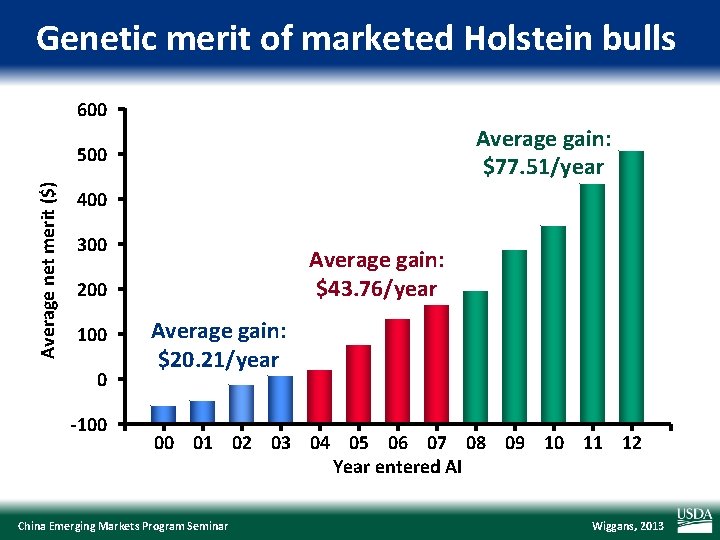
Genetic merit of marketed Holstein bulls 600 Average gain: $77. 51/year Average net merit ($) 500 400 300 Average gain: $43. 76/year 200 100 0 -100 Average gain: $20. 21/year 00 01 02 03 04 05 06 07 08 09 10 11 12 Year entered AI China Emerging Markets Program Seminar Wiggans, 2013

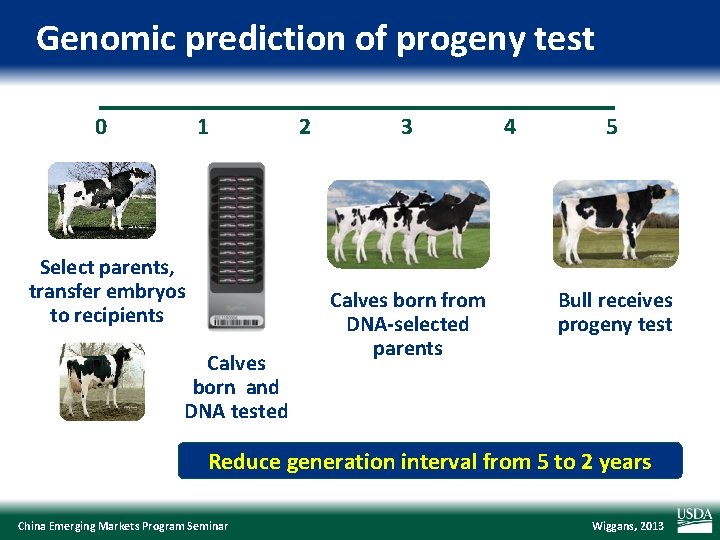
Genomic prediction of progeny test 0 1 Select parents, transfer embryos to recipients Calves born and DNA tested 2 3 Calves born from DNA-selected parents 4 5 Bull receives progeny test Reduce generation interval from 5 to 2 years China Emerging Markets Program Seminar Wiggans, 2013

Benefit of genomics l Determine value of bull at birth l Increase accuracy of selection l Reduce generation interval l Increase selection intensity l Increase rate of genetic gain China Emerging Markets Program Seminar Wiggans, 2013

Why genomics works for dairy cattle l Extensive historical data available l Well-developed genetic evaluation program l Widespread use of AI sires l Progeny-test programs l High-value animals worth the cost of genotyping l Long generation interval that can be reduced substantially by genomics China Emerging Markets Program Seminar Wiggans, 2013

Current organizational roles l Council on Dairy Cattle Breeding (CDCB) responsible for receiving data, computing, and delivering U. S. genetic evaluations for dairy cattle l Animal Improvement Programs Laboratory (AIPL) responsible for research and development to improve the evaluation system l CDCB and USDA employees co-located in Beltsville China Emerging Markets Program Seminar Wiggans, 2013

Funding l l CDCB evaluation calculation and dissemination funded by fee system w Based on animals genotyped w About 80% of revenue from bulls w Higher fees for herds that contribute less information $ AIPL research on evaluation methodology funded by U. S. Federal government China Emerging Markets Program Seminar Wiggans, 2013

Ways to increase accuracy l Automatic addition of traditional evaluations of genotyped bulls when bull is 5 years old l Possible genotyping of 10, 000 bulls with semen in repository l Collaboration with other countries l Use of more SNPs from HD chips l Full sequencing (identify causative mutations) China Emerging Markets Program Seminar Wiggans, 2013

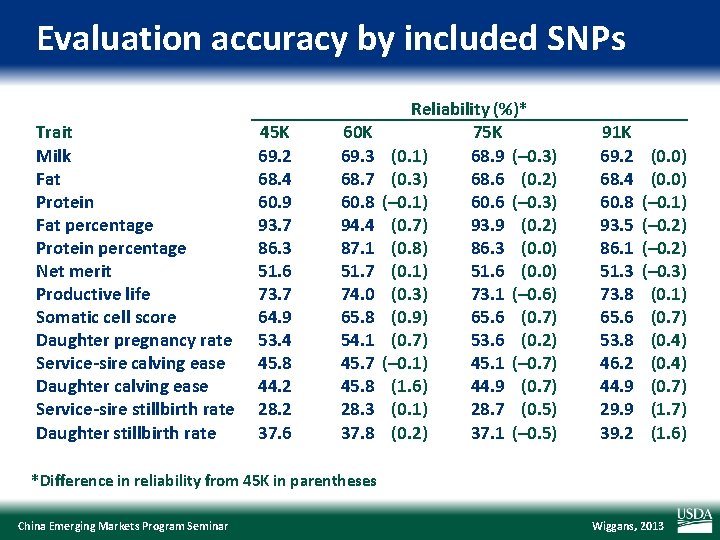
Evaluation accuracy by included SNPs Trait Milk Fat Protein Fat percentage Protein percentage Net merit Productive life Somatic cell score Daughter pregnancy rate Service-sire calving ease Daughter calving ease Service-sire stillbirth rate Daughter stillbirth rate 45 K 69. 2 68. 4 60. 9 93. 7 86. 3 51. 6 73. 7 64. 9 53. 4 45. 8 44. 2 28. 2 37. 6 60 K 69. 3 68. 7 60. 8 94. 4 87. 1 51. 7 74. 0 65. 8 54. 1 45. 7 45. 8 28. 3 37. 8 Reliability (%)* 75 K (0. 1) 68. 9 (– 0. 3) (0. 3) 68. 6 (0. 2) (– 0. 1) 60. 6 (– 0. 3) (0. 7) 93. 9 (0. 2) (0. 8) 86. 3 (0. 0) (0. 1) 51. 6 (0. 0) (0. 3) 73. 1 (– 0. 6) (0. 9) 65. 6 (0. 7) 53. 6 (0. 2) (– 0. 1) 45. 1 (– 0. 7) (1. 6) 44. 9 (0. 7) (0. 1) 28. 7 (0. 5) (0. 2) 37. 1 (– 0. 5) 91 K 69. 2 68. 4 60. 8 93. 5 86. 1 51. 3 73. 8 65. 6 53. 8 46. 2 44. 9 29. 9 39. 2 (0. 0) (– 0. 1) (– 0. 2) (– 0. 3) (0. 1) (0. 7) (0. 4) (0. 7) (1. 6) *Difference in reliability from 45 K in parentheses China Emerging Markets Program Seminar Wiggans, 2013

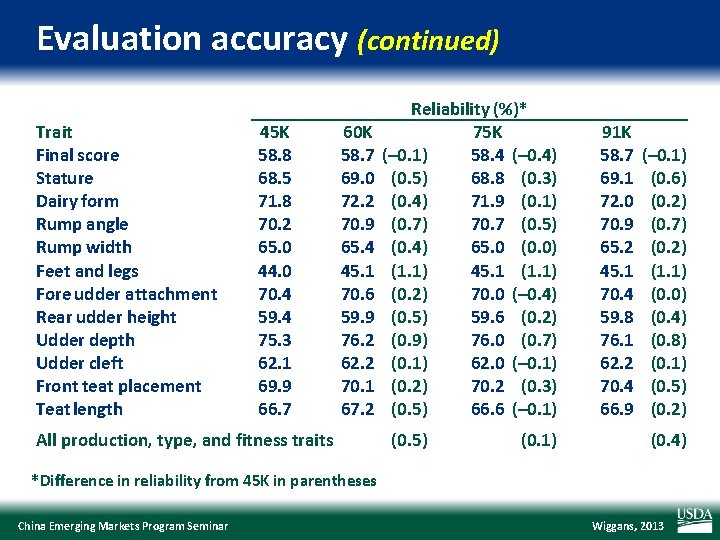
Evaluation accuracy (continued) Trait Final score Stature Dairy form Rump angle Rump width Feet and legs Fore udder attachment Rear udder height Udder depth Udder cleft Front teat placement Teat length 45 K 58. 8 68. 5 71. 8 70. 2 65. 0 44. 0 70. 4 59. 4 75. 3 62. 1 69. 9 66. 7 60 K 58. 7 69. 0 72. 2 70. 9 65. 4 45. 1 70. 6 59. 9 76. 2 62. 2 70. 1 67. 2 All production, type, and fitness traits Reliability (%)* 75 K (– 0. 1) 58. 4 (– 0. 4) (0. 5) 68. 8 (0. 3) (0. 4) 71. 9 (0. 1) (0. 7) 70. 7 (0. 5) (0. 4) 65. 0 (0. 0) (1. 1) 45. 1 (1. 1) (0. 2) 70. 0 (– 0. 4) (0. 5) 59. 6 (0. 2) (0. 9) 76. 0 (0. 7) (0. 1) 62. 0 (– 0. 1) (0. 2) 70. 2 (0. 3) (0. 5) 66. 6 (– 0. 1) (0. 5) (0. 1) 91 K 58. 7 69. 1 72. 0 70. 9 65. 2 45. 1 70. 4 59. 8 76. 1 62. 2 70. 4 66. 9 (– 0. 1) (0. 6) (0. 2) (0. 7) (0. 2) (1. 1) (0. 0) (0. 4) (0. 8) (0. 1) (0. 5) (0. 2) (0. 4) *Difference in reliability from 45 K in parentheses China Emerging Markets Program Seminar Wiggans, 2013

Key issues for the dairy industry l Inbreeding and genetic diversity (including across breeds) l Sequencing, new genes, and mutations l Novel traits, resource populations (feed efficiency, health, milk properties) China Emerging Markets Program Seminar Wiggans, 2013

Application to more traits l Animal’s genotype good for all traits l Traditional evaluations required for accurate estimates of SNP effects l Traditional evaluations not currently available for heat tolerance or feed efficiency l Research populations could provide data for traits that are expensive to measure l Will resulting evaluations work in target population? China Emerging Markets Program Seminar Wiggans, 2013

What’s already planned l Genomic evaluations for new traits w Health (e. g. , resistance to heat stress) w Feed efficiency l Genomic mating programs w Selection of favorable minor alleles w Reduction of genomic inbreeding l Genomic evaluations based on more SNPs (60 K) l Adding SNPs for causative genetic variants China Emerging Markets Program Seminar Wiggans, 2013

What’s already planned (continued) l BARD project (Volcani Center, Israel) w A priori granddaughter design (APGD) w Identification of causative variants for economically important traits l International collaboration on sequencing w United States, United Kingdom, Italy, Canada w Bulls selected using APGD China Emerging Markets Program Seminar Wiggans, 2013

Parentage validation and discovery l Parent-progeny conflicts detected w Animal checked against all other genotypes w Reported to breeds and requesters w Correct sire usually detected l Maternal grandsire (MGS) checking w SNP at a time checking w Haplotype checking more accurate l Breeds moving to accept SNPs in place of microsatellites China Emerging Markets Program Seminar Wiggans, 2013

MGS detection — HAP method l Based on common haplotypes l After imputation of all loci, determine maternal contribution by removing paternal haplotype l Count maternal haplotypes in common with MGS l Remove haplotypes from MGS and check remaining against maternal great-grandsire (MGGS) China Emerging Markets Program Seminar Wiggans, 2013

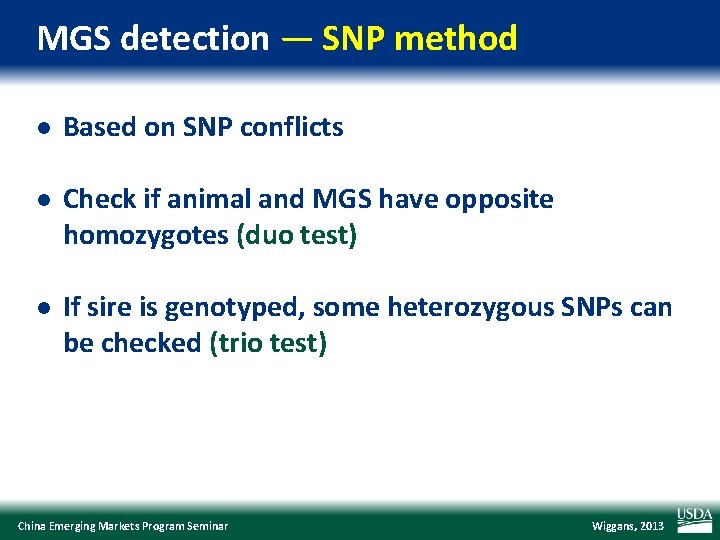
MGS detection — SNP method l Based on SNP conflicts l Check if animal and MGS have opposite homozygotes (duo test) l If sire is genotyped, some heterozygous SNPs can be checked (trio test) China Emerging Markets Program Seminar Wiggans, 2013

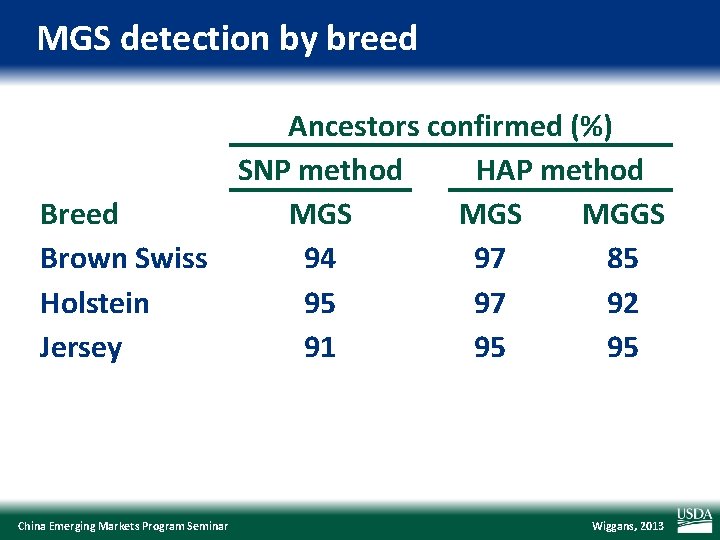
MGS detection by breed Ancestors confirmed (%) SNP method HAP method Breed MGS MGGS Brown Swiss 94 97 85 Holstein 95 97 92 Jersey 91 95 95 China Emerging Markets Program Seminar Wiggans, 2013

Haplotypes affecting fertility l Rapid discovery of new recessive defects w Large numbers of genotyped animals w Affordable DNA sequencing l Determination of haplotype location w Significant number of homozygous animals expected, but none observed w Narrow suspect region with fine mapping w Use sequence data to find causative mutation China Emerging Markets Program Seminar Wiggans, 2013

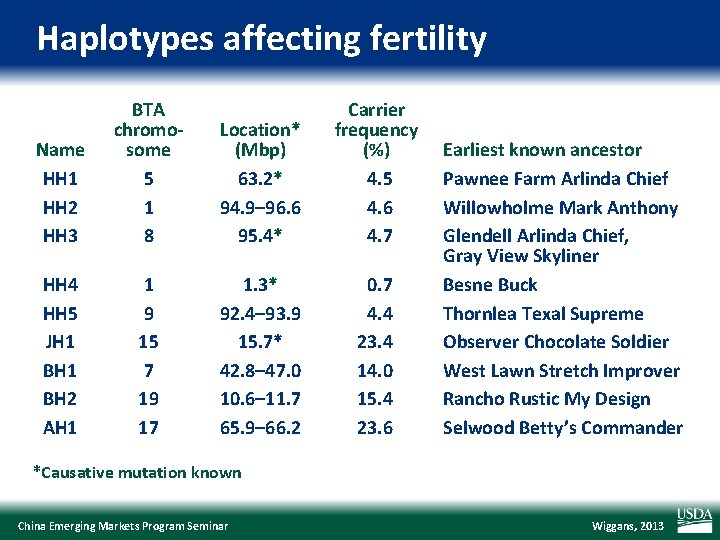
Haplotypes affecting fertility Name HH 1 HH 2 HH 3 BTA chromosome 5 1 8 Location* (Mbp) 63. 2* 94. 9– 96. 6 95. 4* Carrier frequency (%) 4. 5 4. 6 4. 7 HH 4 HH 5 JH 1 BH 2 AH 1 1 9 15 7 19 17 1. 3* 92. 4– 93. 9 15. 7* 42. 8– 47. 0 10. 6– 11. 7 65. 9– 66. 2 0. 7 4. 4 23. 4 14. 0 15. 4 23. 6 Earliest known ancestor Pawnee Farm Arlinda Chief Willowholme Mark Anthony Glendell Arlinda Chief, Gray View Skyliner Besne Buck Thornlea Texal Supreme Observer Chocolate Soldier West Lawn Stretch Improver Rancho Rustic My Design Selwood Betty’s Commander *Causative mutation known China Emerging Markets Program Seminar Wiggans, 2013

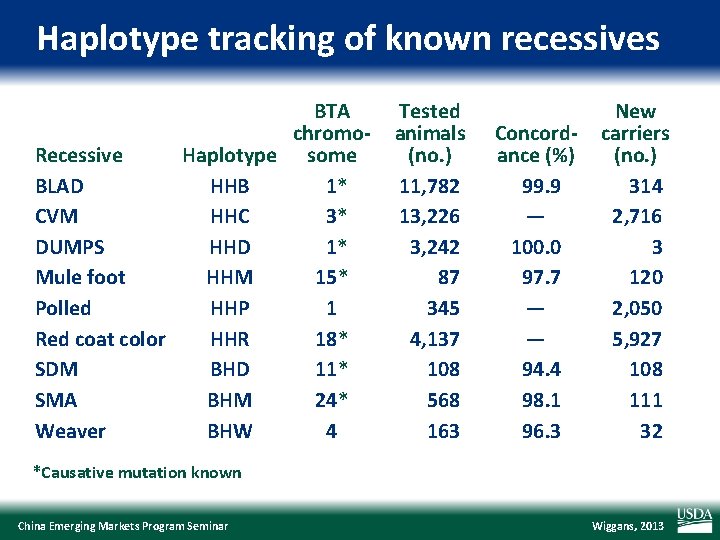
Haplotype tracking of known recessives BTA chromo. Recessive Haplotype some BLAD HHB 1* CVM HHC 3* DUMPS HHD 1* Mule foot HHM 15* Polled HHP 1 Red coat color HHR 18* SDM BHD 11* SMA BHM 24* Weaver BHW 4 Tested animals (no. ) 11, 782 13, 226 3, 242 87 345 4, 137 108 568 163 Concordance (%) 99. 9 — 100. 0 97. 7 — — 94. 4 98. 1 96. 3 New carriers (no. ) 314 2, 716 3 120 2, 050 5, 927 108 111 32 *Causative mutation known China Emerging Markets Program Seminar Wiggans, 2013

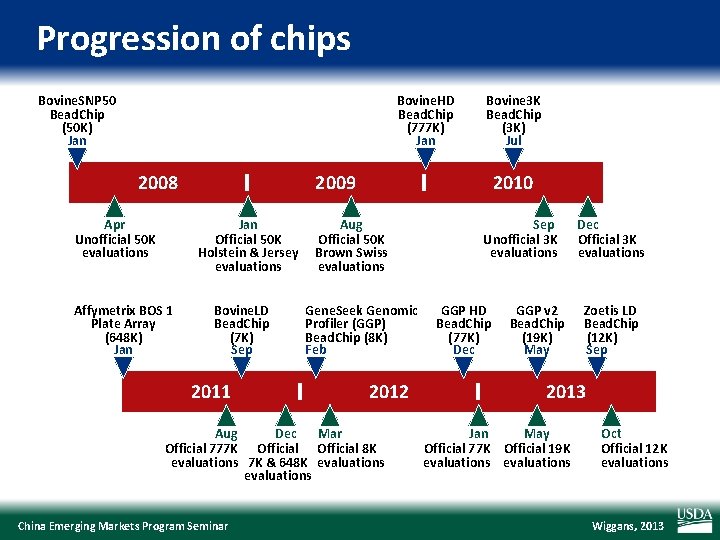
Progression of chips Bovine. SNP 50 Bead. Chip (50 K) Jan Bovine. HD Bead. Chip (777 K) Jan 2008 Apr Unofficial 50 K evaluations 2009 Jan Official 50 K Holstein & Jersey evaluations Affymetrix BOS 1 Plate Array (648 K) Jan Bovine 3 K Bead. Chip (3 K) Jul Bovine. LD Bead. Chip (7 K) Sep 2011 2010 Aug Official 50 K Brown Swiss evaluations Gene. Seek Genomic Profiler (GGP) Bead. Chip (8 K) Feb 2012 Aug Dec Mar Official 777 K Official 8 K evaluations 7 K & 648 K evaluations China Emerging Markets Program Seminar Sep Unofficial 3 K evaluations GGP HD Bead. Chip (77 K) Dec GGP v 2 Bead. Chip (19 K) May Dec Official 3 K evaluations Zoetis LD Bead. Chip (12 K) Sep 2013 Jan May Official 77 K Official 19 K evaluations Oct Official 12 K evaluations Wiggans, 2013

International dairy breeding l Genotype alliances w North America (US, Canada, UK, Italy) w Ireland, New Zealand w Netherlands, Australia w Eurogenomics (Denmark/Sweden/Finland, France, Germany, Netherlands/Belgium, Spain, Poland) l Interbull genomic multitrait across-country evaluation (GMACE) China Emerging Markets Program Seminar Wiggans, 2013

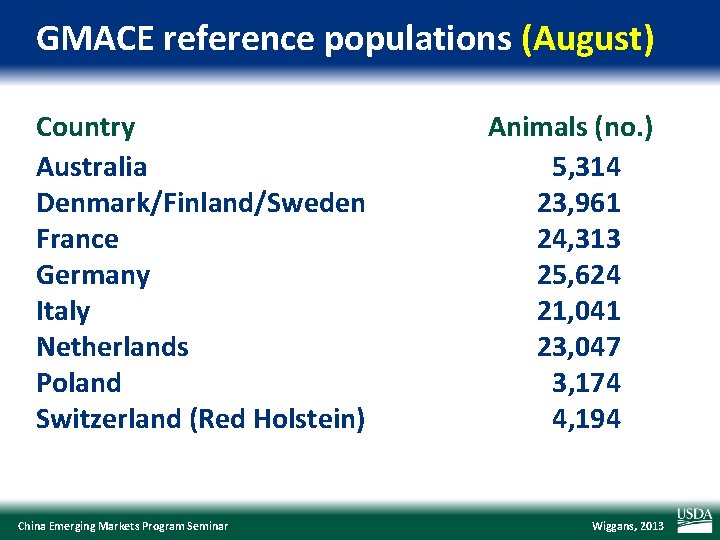
GMACE reference populations (August) Country Australia Denmark/Finland/Sweden France Germany Italy Netherlands Poland Switzerland (Red Holstein) China Emerging Markets Program Seminar Animals (no. ) 5, 314 23, 961 24, 313 25, 624 21, 041 23, 047 3, 174 4, 194 Wiggans, 2013

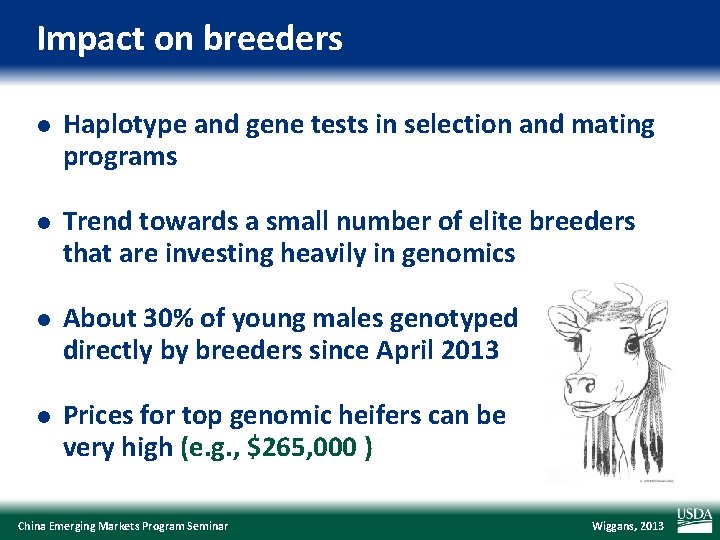
Impact on breeders l Haplotype and gene tests in selection and mating programs l Trend towards a small number of elite breeders that are investing heavily in genomics l About 30% of young males genotyped directly by breeders since April 2013 l Prices for top genomic heifers can be very high (e. g. , $265, 000 ) China Emerging Markets Program Seminar Wiggans, 2013

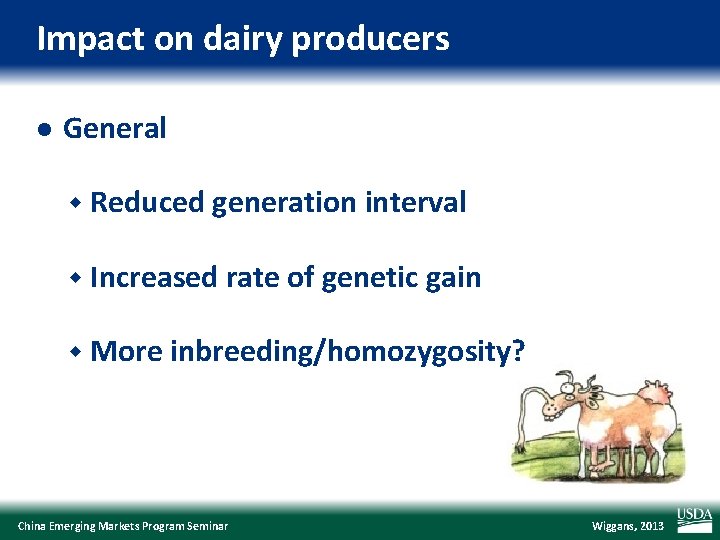
Impact on dairy producers l General w Reduced generation interval w Increased rate of genetic gain w More inbreeding/homozygosity? China Emerging Markets Program Seminar Wiggans, 2013

Impact on dairy producers (continued) l Sires w Higher average genetic merit of available bulls w More rapid increase in genetic merit for all traits w Larger choice of bulls in terms of traits and semen price w Greater use of young bulls China Emerging Markets Program Seminar Wiggans, 2013

Conclusions l Genomic evaluation has dramatically changed dairy cattle breeding l Rate of gain is increasing primarily because of a large reduction in generation interval l Genomic research is ongoing w Detect causative genetic variants w Find more haplotypes affecting fertility w Improve accuracy through more SNPs, more predictor animals, and more traits China Emerging Markets Program Seminar Wiggans, 2013

U. S. genomic evaluation team China Emerging Markets Program Seminar Wiggans, 2013