CTSeq A Gene Sequence Analysis Database Built for

CTSeq : A Gene Sequence Analysis Database Built for Speed (Code-tolerant presentation) September 11, 2001 James E. Ries, M. S. , Depts. CECS & HMI Gordon K. Springer, Ph. D. , Depts. CECS & IATS

Abstract • The discovery and analysis of genomic sequence information generates huge quantities of raw data. To deal with this data in an effective way, we have created a custom database system called CTSeq which is based on Faircom Corporation's CTree database toolkit. This system provides high performance access to a wide variety of sequence information with very low system overhead. CTSeq is modular and general, and should be adaptable to a wide variety of large scale data.

Background (cont. )
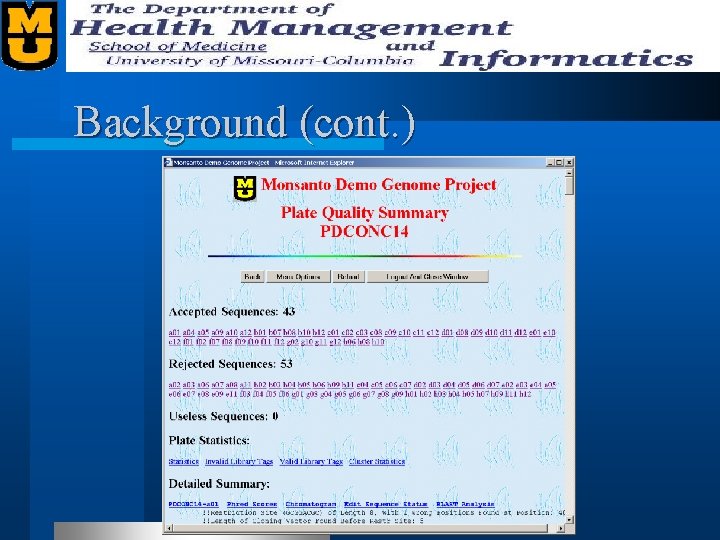
Background (cont. )
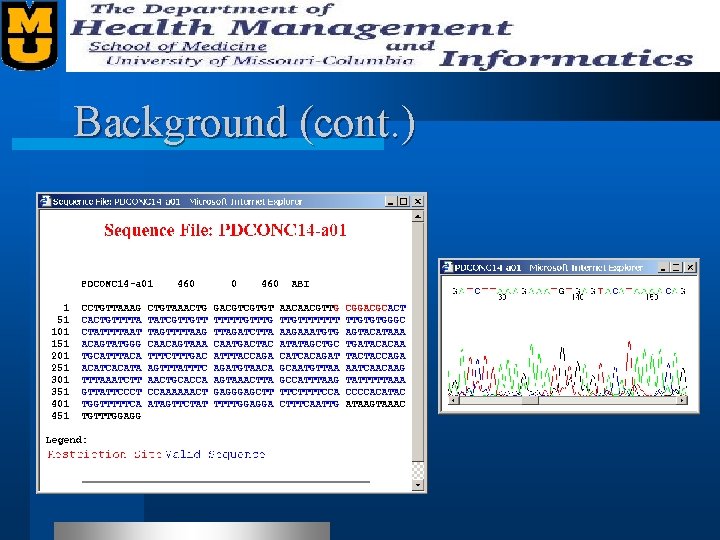
Background (cont. )
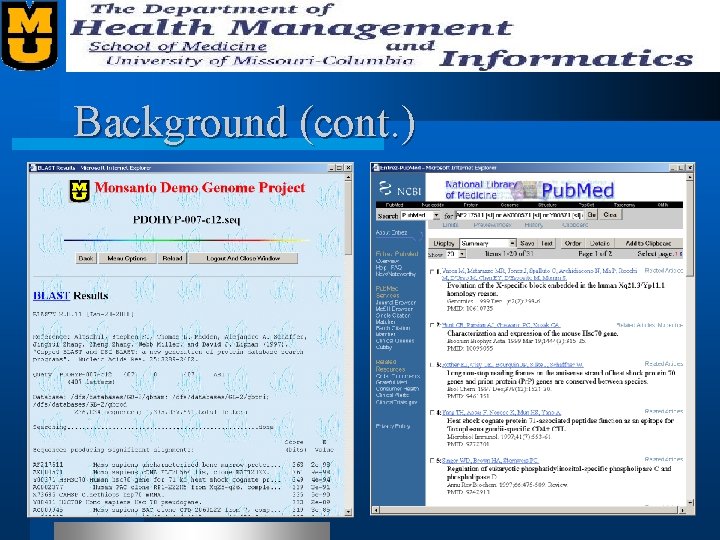
Background (cont. )

Motivation Many current bioinformatics projects are file-based l Monsanto Project Example: l – A single plate (96 wells) generates over 500 data files. – Currently (9/11/2001) approximately 394 plates (500 * 394 = 197, 000 files) – Raw plate data alone for the project takes up over 4 gig of space. – Analysis programs are typically Unix command-line.

Motivation (cont. ) l Many projects run without any “infrastructure” – Researchers must “cut and paste” data from one application to another. – Some data must be transformed in format in order to work in another application, and this is difficult if not impossible for many researchers.

Motivation (cont. ) l Even powerful machines can be “slow” in processing this enormous amount of data. – Darwin is a Compaq True 64 Unix box with 4 processors and 2 Gig (4 Gig? ) of RAM – Darwin takes approximately 1 week to completely search Genbank for Monsanto project sequences
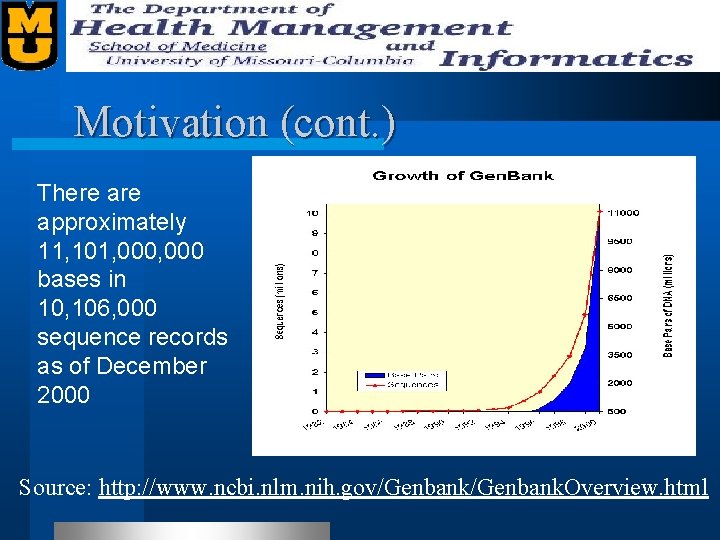
Motivation (cont. ) There approximately 11, 101, 000 bases in 10, 106, 000 sequence records as of December 2000 Source: http: //www. ncbi. nlm. nih. gov/Genbank. Overview. html

Solution Discussion l Our solution is to build a custom DBMS using Faircom’s ISAM toolkit (C-Tree+). – Allows multiple interfaces (C, C++, ODBC, CGI, etc. ) – Provides complete control of important implementation details (for performance) – Avoids costly and less effective “shrinkwrapped” solutions l Our DBMS (CTSeq) ties together all bioinformatic data and internally does necessary transformations.
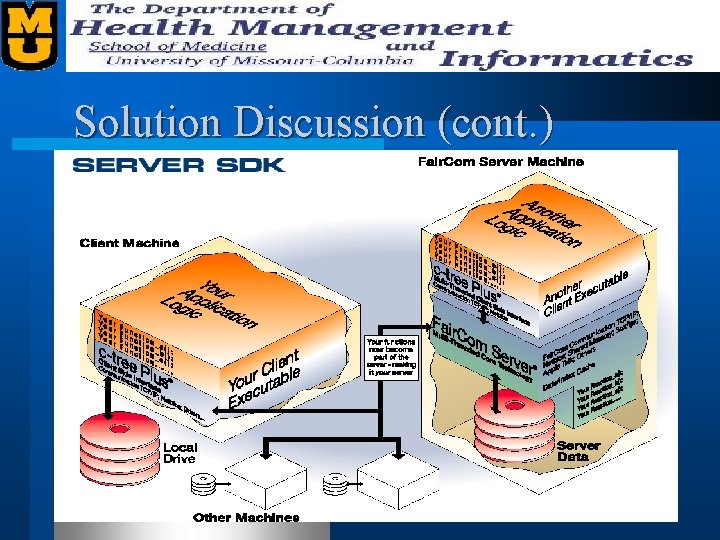
Solution Discussion (cont. )
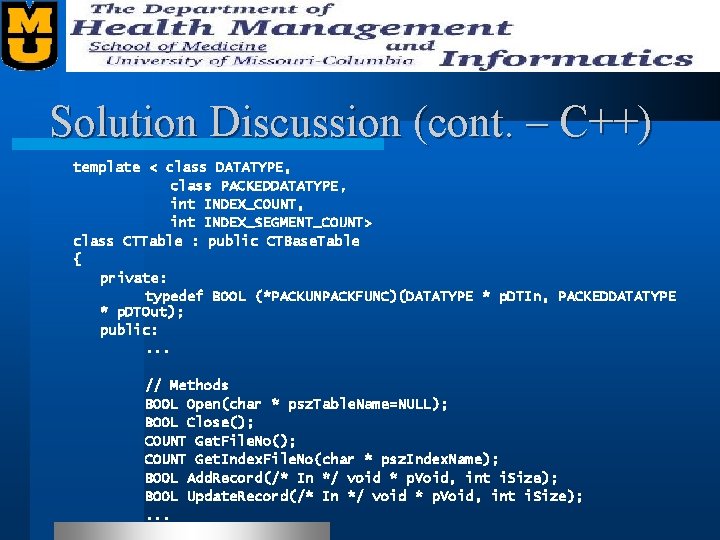
Solution Discussion (cont. – C++) template < class DATATYPE, class PACKEDDATATYPE, int INDEX_COUNT, int INDEX_SEGMENT_COUNT> class CTTable : public CTBase. Table { private: typedef BOOL (*PACKUNPACKFUNC)(DATATYPE * p. DTIn, PACKEDDATATYPE * p. DTOut); public: . . . // Methods BOOL Open(char * psz. Table. Name=NULL); BOOL Close(); COUNT Get. File. No(); COUNT Get. Index. File. No(char * psz. Index. Name); BOOL Add. Record(/* In */ void * p. Void, int i. Size); BOOL Update. Record(/* In */ void * p. Void, int i. Size); . . .

Solution Discussion (cont. – C) CEXTERN BOOL Init. CTDB(); CEXTERN BOOL Un. Init. CTDB(); CEXTERN BOOL Open. CTTable( /* In */ TABLETYPE tt, /* Out */ CTDBHANDLE * ph. DB); CEXTERN BOOL Open. CTTable. By. Name( /* In */ TABLETYPE tt, /* Out */ CTDBHANDLE * ph. DB, /* In */ char * psz. DBName); CEXTERN BOOL Close. CTDB(/* In */ CTDBHANDLE h. DB); CEXTERN BOOL Find. First. CTRecord( /* In */ CTDBHANDLE h. DB, /* Out */ void * p. Seq); CEXTERN BOOL Find. Next. CTRecord( /* In */ CTDBHANDLE h. DB, /* Out */ void * p. Seq); CEXTERN BOOL Add. CTRecord( /* In */ CTDBHANDLE h. DB, /* In */ void * p. Seq, /* In */ int i. Record. Size); . . .

Solution Discussion (cont. ) typedef struct _SEQUENCE { TEXT int int int char TEXT unsigned char } SEQUENCE, * PSEQUENCE; sz. Plate. Name[MAX_PLATE_NAME]; sz. Well. Name[MAX_WELL_NAME]; i. Seq. Start; i. Seq. End; i. Restriction. Start; i. Restriction. End; i. Lib. Tag. Start; i. Lib. Tag. End; i. Poly. AStart; i. Poly. AEnd; i. Poly. ASignal. Start; i. Poly. ASignal. End; i. Vector. Start; i. Vector. End; c. Status; sz. Bases[MAX_BASES]; u. Phred. Scores[MAX_BASES];

Solution Discussion (cont. ) //////////////////// // CHROMATOGRAM Table //////////////////// typedef struct _CHROMATOGRAM { TEXT sz. Plate. Name[MAX_PLATE_NAME]; TEXT sz. Well. Name[MAX_WELL_NAME]; int i. Chromatogram. Size; unsigned char blob. Chromatogram[MAX_CHROM_SIZE]; } CHROMATOGRAM, * PCHROMATOGRAM;

Solution Discussion (cont. ) //////////////////// // TRANSACTION Table //////////////////// typedef struct _TRANSACTION { TEXT sz. Plate. Name[MAX_PLATE_NAME]; TEXT sz. Well. Name[MAX_WELL_NAME]; TEXT sz. User. ID[MAX_USER_ID]; int i. Seq. Start; int i. Seq. End; time_t time. Stamp; char c. Status; TEXT sz. Comment[MAX_COMMENT]; } TRANSACTION, * PTRANSACTION;
![Solution Discussion (cont. ) typedef struct _CLUSTER { TEXT sz. Plate. Name[MAX_PLATE_NAME]; TEXT sz. Solution Discussion (cont. ) typedef struct _CLUSTER { TEXT sz. Plate. Name[MAX_PLATE_NAME]; TEXT sz.](http://slidetodoc.com/presentation_image_h2/be4bd06fe2dfeb26fa5a36476e7f7b7f/image-18.jpg)
Solution Discussion (cont. ) typedef struct _CLUSTER { TEXT sz. Plate. Name[MAX_PLATE_NAME]; TEXT sz. Well. Name[MAX_WELL_NAME]; TEXT sz. Cluster. Class. Name[MAX_CLUSTER_NAME]; // e. g. , "Folicle" for a "T" type cluster unsigned int u. Cluster. Number; unsigned char b. Seq. Type; // Primary - P; Secondary - S unsigned char b. Cluster. Type; // Plate - P; Library - L; // Tissue - T; Unigene (project) - U } CLUSTER, * PCLUSTER;

Solution Discussion (cont. ) typedef struct _BLAST { TEXT TEXT } BLAST, * PBLAST; sz. Plate. Name[MAX_PLATE_NAME]; sz. Well. Name[MAX_WELL_NAME]; sz. Source[MAX_SOURCE]; sz. GI[MAX_GI]; sz. Accession[MAX_ACCESSION]; sz. Locus[MAX_LOCUS]; sz. Annotation[MAX_ANNOTATION];

Solution Discussion (cont. ) static inline CTSequence. Table * New. CTSequence. Table() { const int i. Seg. Off. Sets []={0, MAX_PLATE_NAME+MAX_WELL_NAME}; . . . return (new CTSequence. Table("Sequence", SEQUENCE_FIXED, USHRT_MAX, SHARED | VLENGTH, SHARED | ct. FIXED, i. Seg. Off. Sets, i. Seg. Lengths, i. Seg. Modes, i. Key. Lens, i. Key. Types, i. Key. Allow. Dups, i. Key. Null. Checks, . . .

Solution Discussion (cont. ) // A Simple example of using CTSeq library Init. CTDB() if (Open. CTTable. By. Name(SEQUENCE_TABLE, &h. DB, psz. Sequence. Table)) { Str. Upper(sz. Plate. Name); f. RC=First. Len. Field. Query(h. DB, "Plate. Name", sz. Plate. Name, sizeof(sz. Plate. Name), &seq); while (f. RC) { printf("%s-%s : start=%d, end=%dn", seq. sz. Plate. Name, seq. sz. Well. Name, seq. i. Seq. Start, seq. i. Seq. End); f. RC=Next. Field. Query(h. DB, &seq); } Close. CTDB(h. DB); } Un. Init. CTDB();
Solution Discussion (cont. )
Solution Discussion (cont. )

Benchmarks l We benchmarked our “Sequence Quality” table. – 27, 456 records – Records contain • • • Plate Name Well Name Bases Various offsets (restriction site, lib tag, etc. ) Status Quality Scores (Phred)
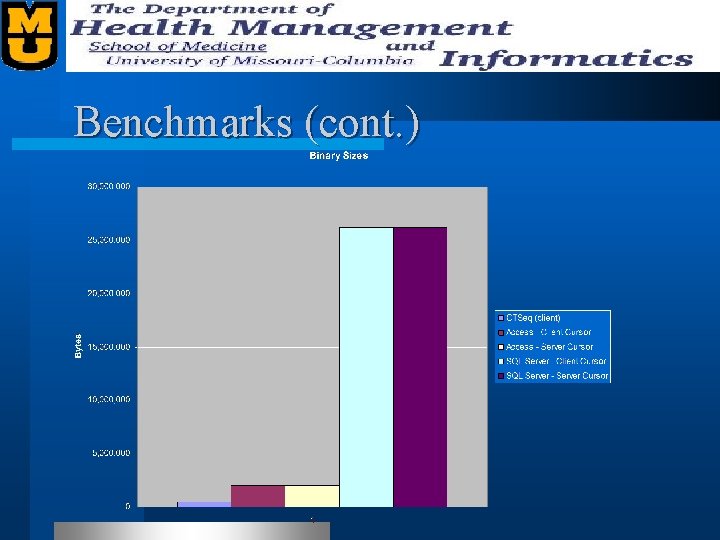
Benchmarks (cont. )
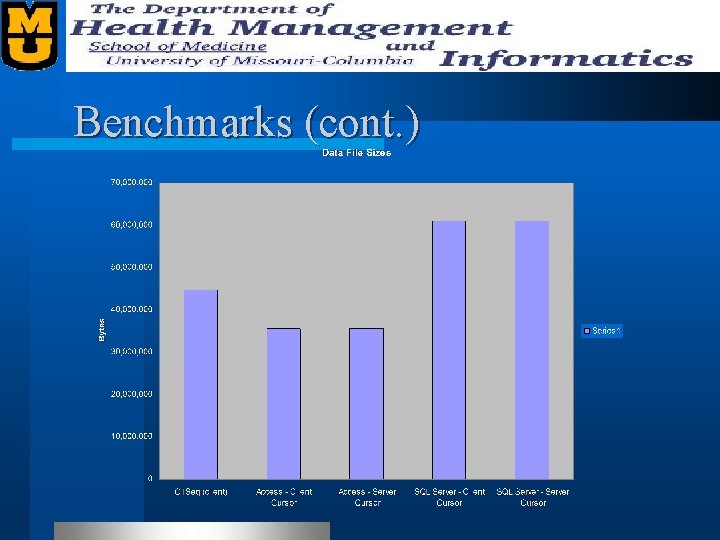
Benchmarks (cont. )
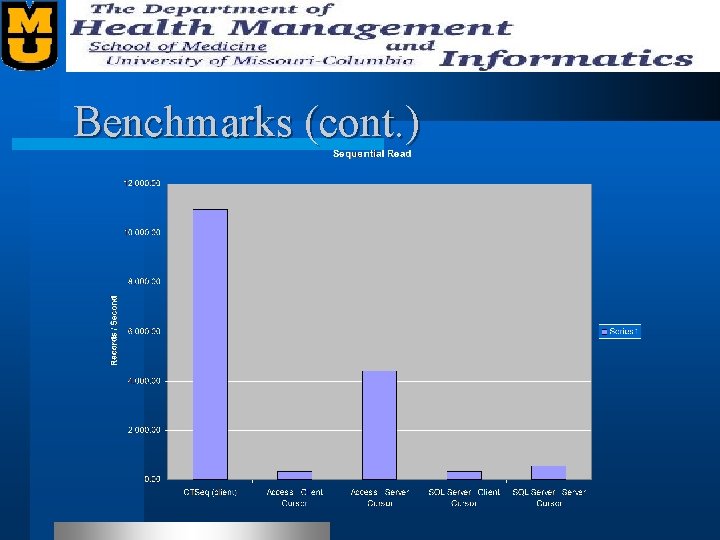
Benchmarks (cont. )
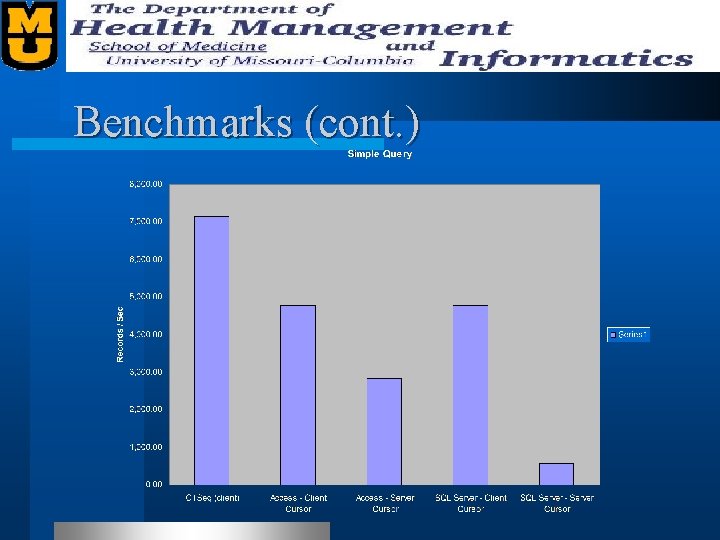
Benchmarks (cont. )
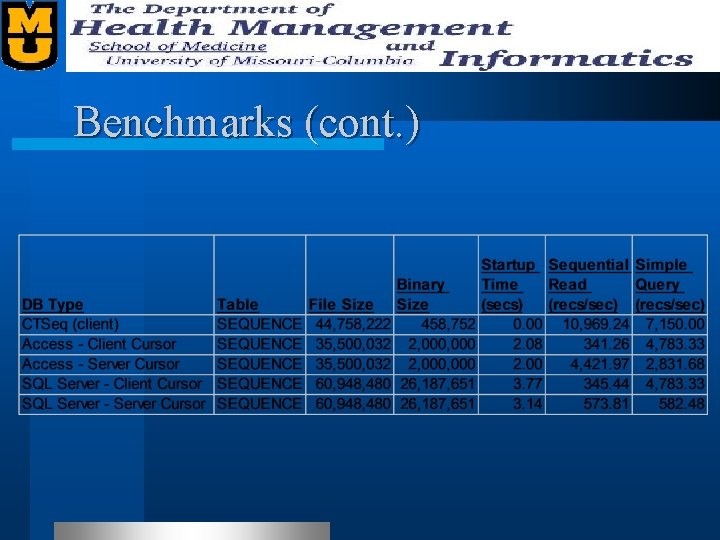
Benchmarks (cont. )

A Straw Man l Isn’t this just another case of “Not Invented Here” foolish pride? – “Standard” solutions such as Microsoft SQL Server or Oracle generally don’t provide the control needed for a truly performance-bound project. – Server-side extensions which would be necessary if using “standard” solutions would be just as “proprietary” as CTree. – Oracle or MS SQL Server typically require a full-time administrator and are financially expensive in general.

A Straw Man (cont. ) l However, “off-the-shelf” DBMS solutions have advantages – Existing support infrastructure – Flexibility – Mature development tools l So, a custom DBMS is really only needed where truly high performance is required.

We’ve only just begun… l Currently, only “core” data is in our DB – – – l Have trimming/quality information Have chromatograms (binary blobs) Adding BLAST results Adding cluster information Adding transaction history Plan to integrate our security system which currently uses another DBMS Switch to client-server (currently using “stand-alone” compile)

We’ve only just begun…(cont. ) l Server-side processing modules – BLAST – MSA – GCG? l Automatic compression / decompression l Automatic encryption / decryption l ODBC Driver installation/test

Acknowledgements l Faircom Corporation l Monsanto l National Company Library of Medicine

Questions • http: //swine. rnet. missouri. edu/Demo/index. html • http: //jimries. com/Seq. CTree/ • Jim. R@acm. org
- Slides: 35