Crystallographic structure analysis of Chitinase enzyme from Corms





- Slides: 25

Crystallographic structure analysis of Chitinase enzyme from Corms of Crocus vernus Dr. Ahmed Akrem Bahauddin Zakariya University, Multan 29. 04. 14 1

Importance of Chitinase Ø Chitinases catalyze the hydrolysis of chitin 1 Ø Chitinases occur in a wide range of organisms, including plants, animals, viruses, bacteria, fungi and insects, and play a variety of roles in these organisms. 2 Ø Plant chitinases are a structurally diverse group with respect to their physical properties, enzymatic activities and localization. 3 Ø Chitin is an unbranched homopolymer of 1, 4 -linked N-acetyl-d-glucosamine. 4 Ø Chitin is not a component of mammalian cells; it occurs widely elsewhere in nature and is abundant in human pathogens. Ø More than 75% of the industrial enzymes are hydrolases. 5 1 Bishop 2 et al. , 2000; 2 Brunner et al. , 1998; 2 Hoell et al. , 2005; 3 Collinge et al. , 1993; 4 Butt & Sultan, 2010; 5 Leishola et al. , 2005

Aims and Objectives 1. Isolation and purification of the chitinase protein from C. vernus 2. Crystallization of the purified protein 3. X-ray diffraction data collection Ø 3 D molecular structure determination 3

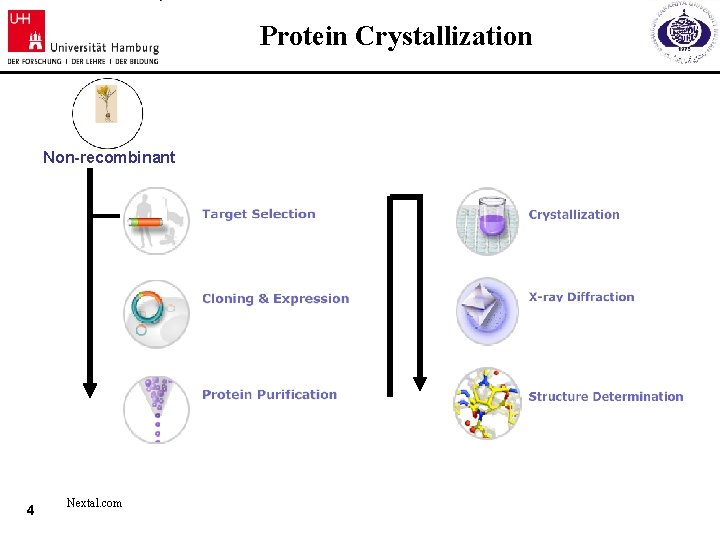
Protein Crystallization Non-recombinant 4 Nextal. com

Crocus vernus Ø Genus Crocus belongs to family Iridaceae. Ø A perennial flowering plant found in Central and Southern Europe, North Africa, Middle East, Central Asia to China. Ø Most expansive spice “Saffron” is from Crocus sativus L. Ø Chitinases & Lectins are ´´ Defense-related plant proteins``. Ø Plant chitinases are a structurally diverse group. 3 Ø So far no crystal structure of this plant is deposited in the Protein Data Bank. 5 3 Collinge et al. , 1993,

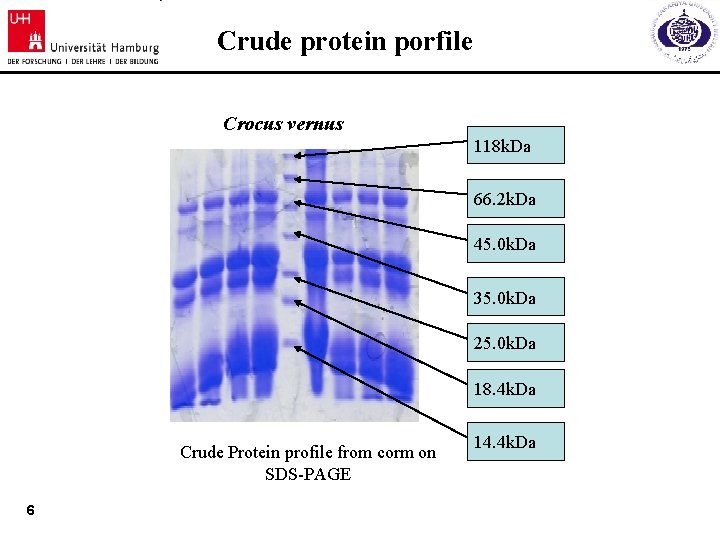
Crude protein porfile Crocus vernus 118 k. Da 66. 2 k. Da 45. 0 k. Da 35. 0 k. Da 25. 0 k. Da 18. 4 k. Da Crude Protein profile from corm on SDS-PAGE 6 14. 4 k. Da

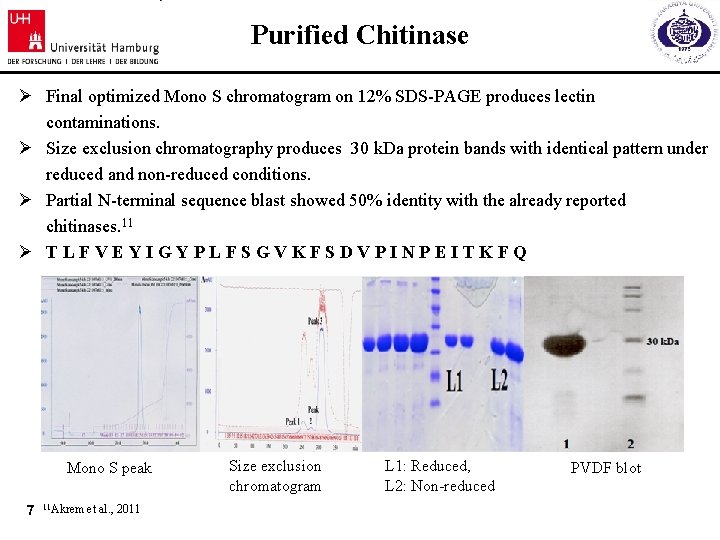
Purified Chitinase Ø Final optimized Mono S chromatogram on 12% SDS-PAGE produces lectin contaminations. Ø Size exclusion chromatography produces 30 k. Da protein bands with identical pattern under reduced and non-reduced conditions. Ø Partial N-terminal sequence blast showed 50% identity with the already reported chitinases. 11 Ø TLFVEYIGYPLFSGVKFSDVPINPEITKFQ Mono S peak 7 11 Akrem et al. , 2011 Size exclusion chromatogram L 1: Reduced, L 2: Non-reduced PVDF blot

Purification techniques/Instruments Ø Protein characterization • N-terminal amino acid sequencing • MALDI/TOF Mass spectrometry • SDS-PAGE Ø Protein Purification • Ammonium sulfate precipitation • Dialysis • Column Chromatography • Gel filtration • Ion exchanger columns (Cation/Anion: Isoelectric p. H or p. I) 8

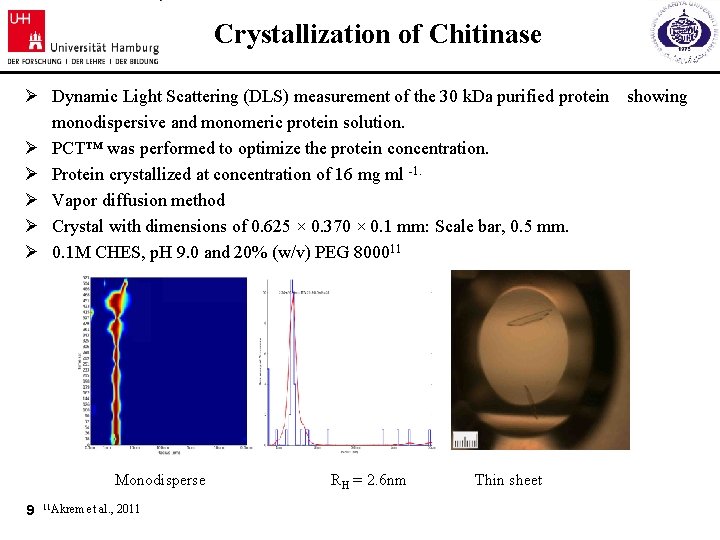
Crystallization of Chitinase Ø Dynamic Light Scattering (DLS) measurement of the 30 k. Da purified protein showing monodispersive and monomeric protein solution. Ø PCT™ was performed to optimize the protein concentration. Ø Protein crystallized at concentration of 16 mg ml -1. Ø Vapor diffusion method Ø Crystal with dimensions of 0. 625 × 0. 370 × 0. 1 mm: Scale bar, 0. 5 mm. Ø 0. 1 M CHES, p. H 9. 0 and 20% (w/v) PEG 800011 Monodisperse 9 11 Akrem et al. , 2011 RH = 2. 6 nm Thin sheet

Vapor Diffusion Methods Hanging Drop 10 Sitting Drop

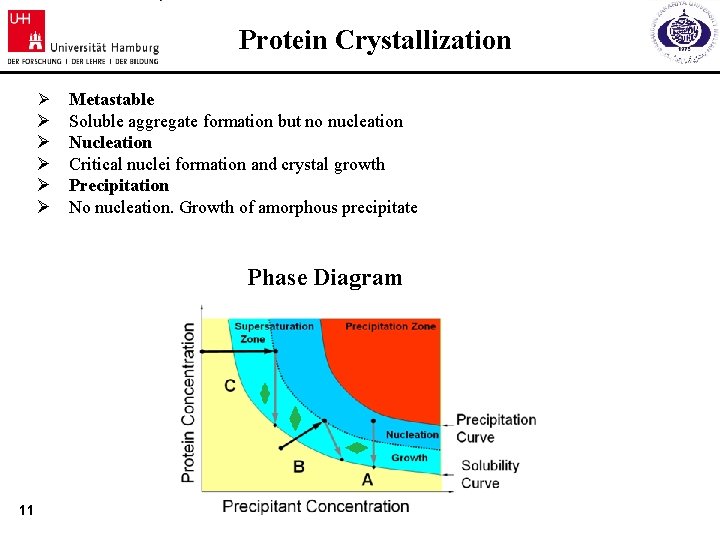
Protein Crystallization Ø Ø Ø Metastable Soluble aggregate formation but no nucleation Nucleation Critical nuclei formation and crystal growth Precipitation No nucleation. Growth of amorphous precipitate Phase Diagram 11

Crystallization Machinery Nanodrop (Protein quantification) Zinsser Pipetting Robot (Digilab Genomic Solution, Germany) 12 Dynamic Light Scattering (Monodispersity) UV-microscope

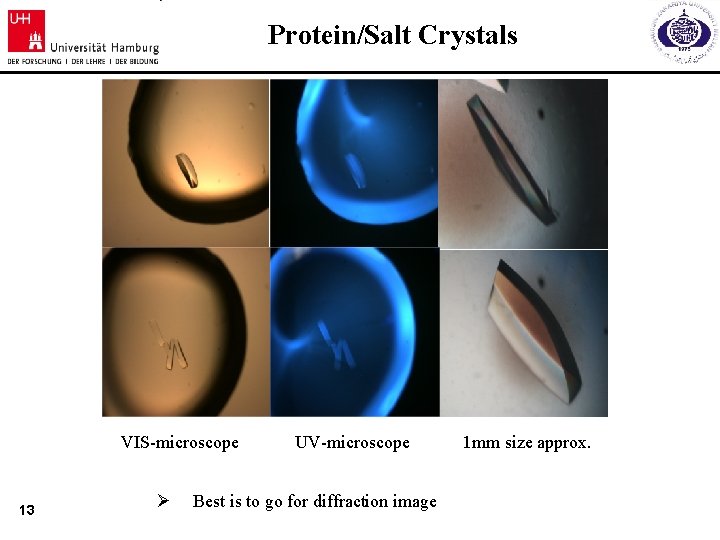
Protein/Salt Crystals VIS-microscope 13 Ø UV-microscope Best is to go for diffraction image 1 mm size approx.

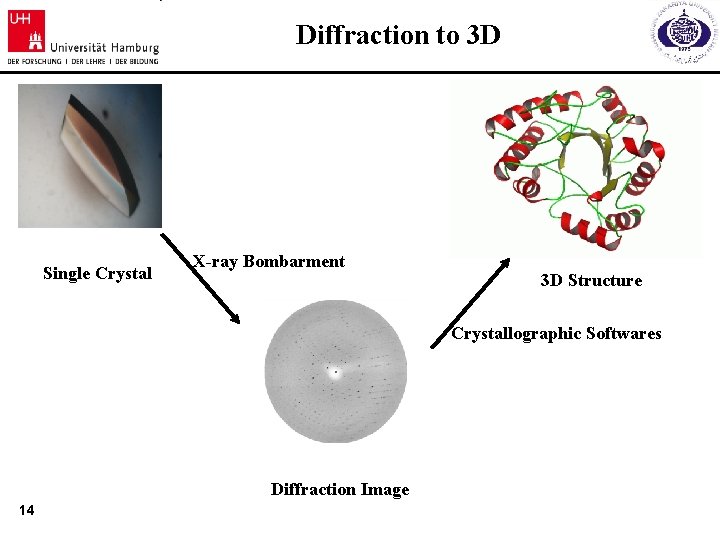
Diffraction to 3 D Single Crystal X-ray Bombarment 3 D Structure Crystallographic Softwares Diffraction Image 14

X-ray Diffraction Data Ø Ø Diffractometer Rotating anode Synchrotron: the best ultimate choice A synchrotron is a particular type of cyclic particle accelerator in which the magnetic field (to turn the particles so they circulate) and the electric field (to accelerate the particles) are carefully synchronized with the travelling particle beam Ø Deutsches Elecktronen Synchrotron (DESY), Hamburg, Germany Ø Approx. 1000 scientists from more than 30 countries around the world are working (2008) Ø Few countries in the world are enjoying this facility 15 Diamond, UK ESRF, France DESY, Germany

Crystal Mounting Ø Nylon loops to fish out crystals Ø Goniohead Ø X-ray gun Ø Cryonozzle Ø Microscope Ø Beamstop Ø Detectors 16

Bioinformatics Ø Imosflm; Scala Ø Denzo; Scalepack Ø CCP 4 i Suite Ø Molecular Replacment; Molrep, Phaser, Mrbump Ø Homer Ø COOT Ø Refmac 5 Ø Protein Data Bank Ø Pdbsum Ø Pdb goodies Ø Chimera Ø Pymol Ø Auto-Rickshaw 17

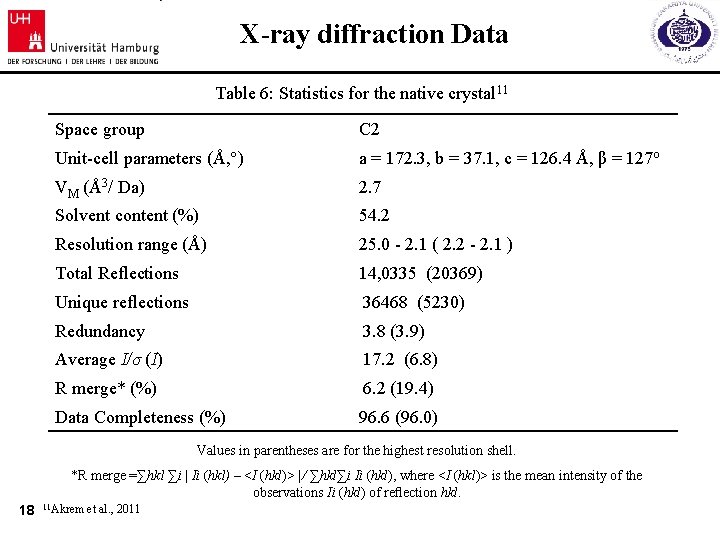
X-ray diffraction Data Table 6: Statistics for the native crystal 11 Space group C 2 Unit-cell parameters (Å, °) a = 172. 3, b = 37. 1, c = 126. 4 Å, β = 127 o VM (Å3/ Da) 2. 7 Solvent content (%) 54. 2 Resolution range (Å) 25. 0 - 2. 1 ( 2. 2 - 2. 1 ) Total Reflections 14, 0335 (20369) Unique reflections 36468 (5230) Redundancy 3. 8 (3. 9) Average I/σ (I) 17. 2 (6. 8) R merge* (%) 6. 2 (19. 4) Data Completeness (%) 96. 6 (96. 0) Values in parentheses are for the highest resolution shell. *R merge =∑hkl ∑i | Ii (hkl) – <I (hkl)> | ⁄ ∑hkl∑i Ii (hkl), where <I (hkl)> is the mean intensity of the observations Ii (hkl) of reflection hkl. 18 11 Akrem et al. , 2011

mtz and pdb files Ø Out put of first processing is a single mtz file of few MB Ø Electron density map Ø Second important file is pdb file based on sequence homology from Protein Data Bank (PDB) like INAR for Narbonin vicia Ø MR strategy to solve the phase information Ø Sequence identity of atleast 40% Ø Clustalw 2, www. pdb. org Ø Pdbgoodies input page Ø Phase information and Coordinate information 19

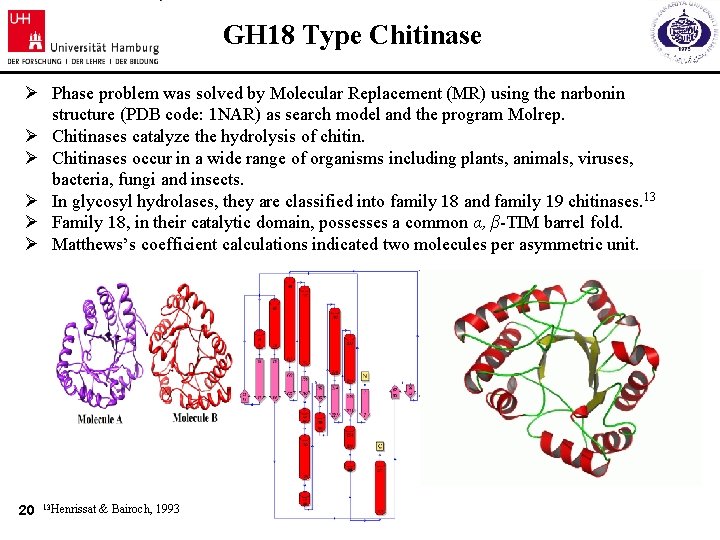
GH 18 Type Chitinase Ø Phase problem was solved by Molecular Replacement (MR) using the narbonin structure (PDB code: 1 NAR) as search model and the program Molrep. Ø Chitinases catalyze the hydrolysis of chitin. Ø Chitinases occur in a wide range of organisms including plants, animals, viruses, bacteria, fungi and insects. Ø In glycosyl hydrolases, they are classified into family 18 and family 19 chitinases. 13 Ø Family 18, in their catalytic domain, possesses a common α, β-TIM barrel fold. Ø Matthews’s coefficient calculations indicated two molecules per asymmetric unit. 20 13 Henrissat & Bairoch, 1993

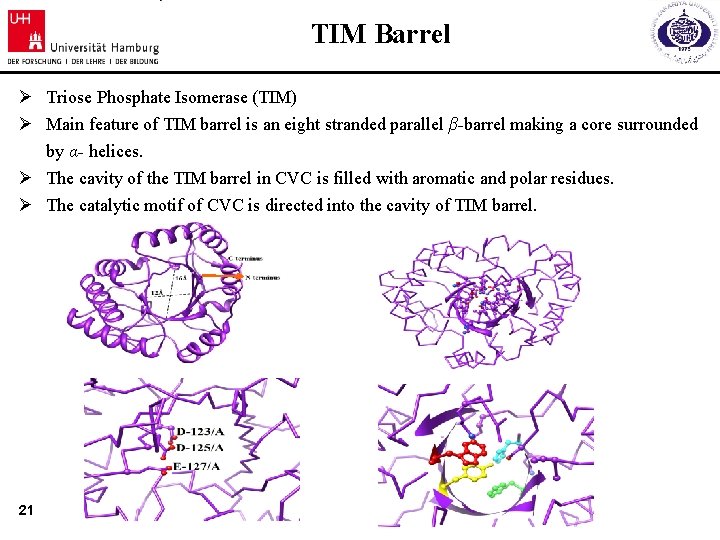
TIM Barrel Ø Triose Phosphate Isomerase (TIM) Ø Main feature of TIM barrel is an eight stranded parallel β-barrel making a core surrounded by α- helices. Ø The cavity of the TIM barrel in CVC is filled with aromatic and polar residues. Ø The catalytic motif of CVC is directed into the cavity of TIM barrel. 21

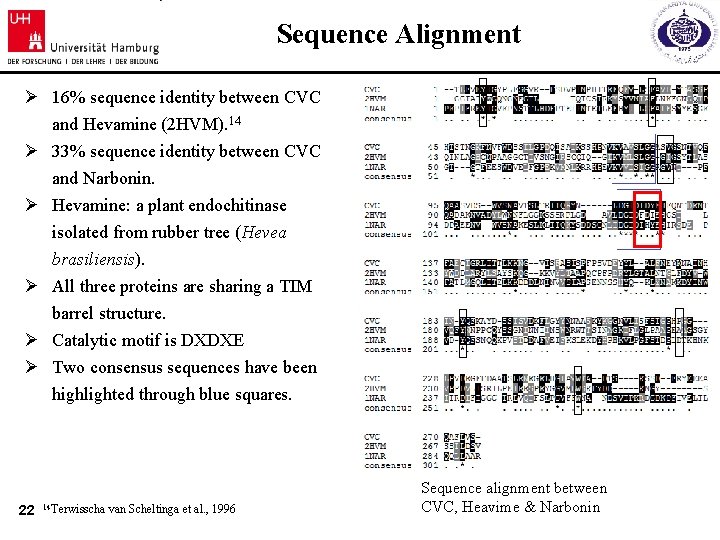
Sequence Alignment Ø 16% sequence identity between CVC and Hevamine (2 HVM). 14 Ø 33% sequence identity between CVC and Narbonin. Ø Hevamine: a plant endochitinase isolated from rubber tree (Hevea brasiliensis). Ø All three proteins are sharing a TIM barrel structure. Ø Catalytic motif is DXDXE Ø Two consensus sequences have been highlighted through blue squares. 22 14 Terwisscha van Scheltinga et al. , 1996 Sequence alignment between CVC, Heavime & Narbonin

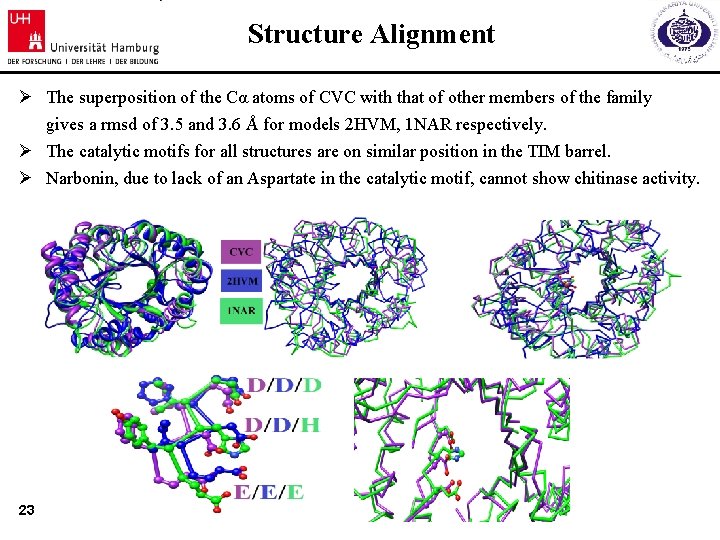
Structure Alignment Ø The superposition of the Cα atoms of CVC with that of other members of the family gives a rmsd of 3. 5 and 3. 6 Å for models 2 HVM, 1 NAR respectively. Ø The catalytic motifs for all structures are on similar position in the TIM barrel. Ø Narbonin, due to lack of an Aspartate in the catalytic motif, cannot show chitinase activity. 23

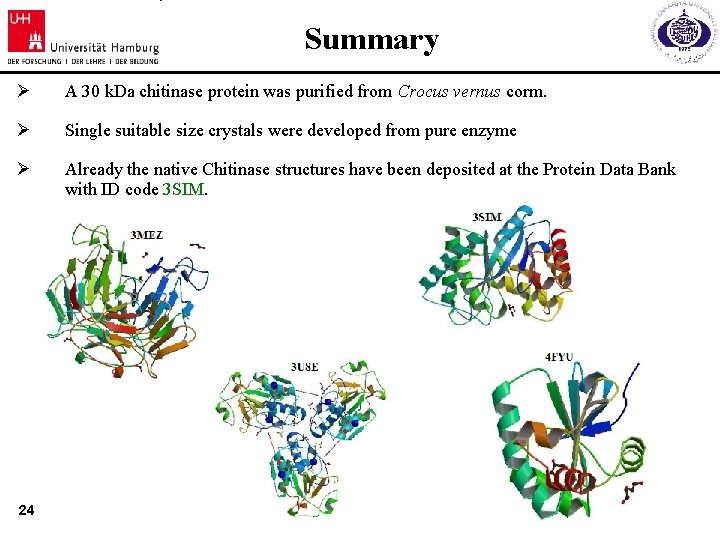
Summary Ø A 30 k. Da chitinase protein was purified from Crocus vernus corm. Ø Single suitable size crystals were developed from pure enzyme Ø Already the native Chitinase structures have been deposited at the Protein Data Bank with ID code 3 SIM. 24

Thanks for kind attention 25