CrossSpecies MultiScalar Resolution of the Developing Lung Transcriptome


- Slides: 7

Cross-Species Multi-Scalar Resolution of the Developing Lung Transcriptome Alvin. T. Kho 1*, Soumyaroop Bhattacharya 3*, Vincent J. Carey 3, Kelan G. Tantisira 3, Roger Gaedigk 2, Scott T. Weiss 3, J. Steven Leeder 2 and Thomas J. Mariani 3. 1 Children’s Hospital Informatics Program, Children’s Hospital Boston, MA, 2 Department of Pediatrics, Children’s Mercy Hospital, Kansas City, MO, and 3 Department of Medicine and the Channing Laboratory, Brigham and Women’s Hospital, Boston, MA. Supported by NIH Grants: HL 71885 (TJ Mariani); HL 65899 (ST Weiss)

Introduction • Regulatory mechanisms of human lung development are poorly defined • Animal models complement human studies • Can we resolve human developmental gene expression patterns using mouse data?

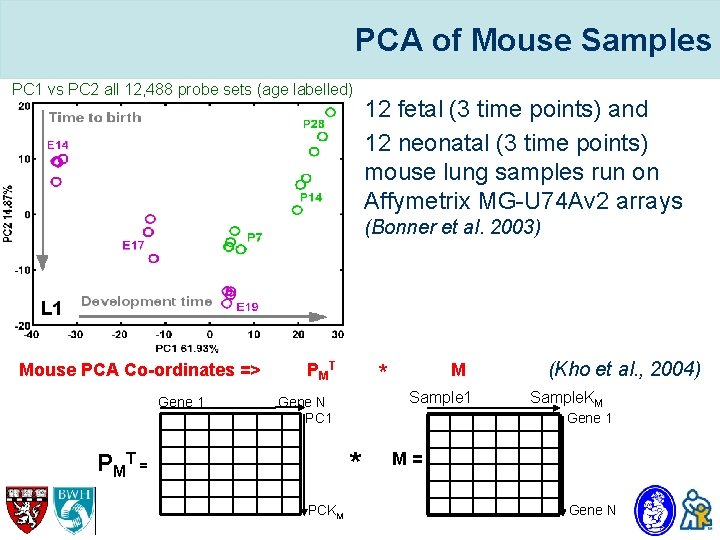
PCA of Mouse Samples PC 1 vs PC 2 all 12, 488 probe sets (age labelled) • 12 fetal (3 time points) and • 12 neonatal (3 time points) mouse lung samples run on Affymetrix MG-U 74 Av 2 arrays (Bonner et al. 2003) Mouse PCA Co-ordinates => Gene 1 PMT Sample 1 Gene N PC 1 PCKM (Kho et al. , 2004) Sample. KM Gene 1 * P MT = M * M= Gene N

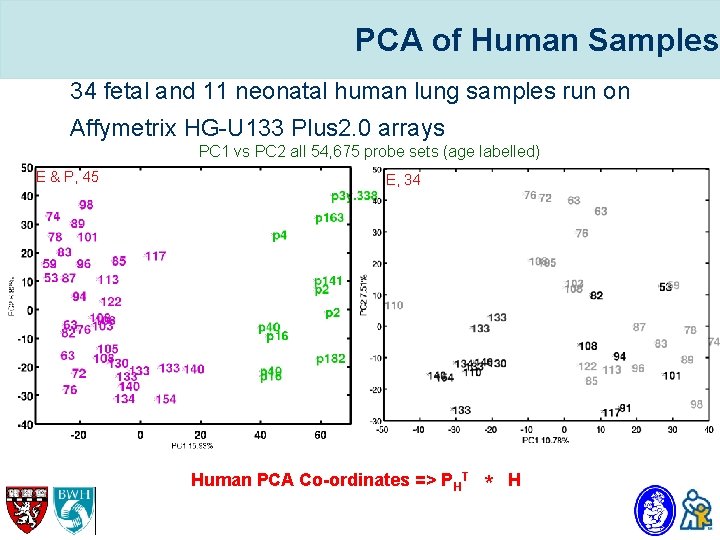
PCA of Human Samples • 34 fetal and 11 neonatal human lung samples run on Affymetrix HG-U 133 Plus 2. 0 arrays PC 1 vs PC 2 all 54, 675 probe sets (age labelled) E & P, 45 E, 34 Human PCA Co-ordinates => PHT * H

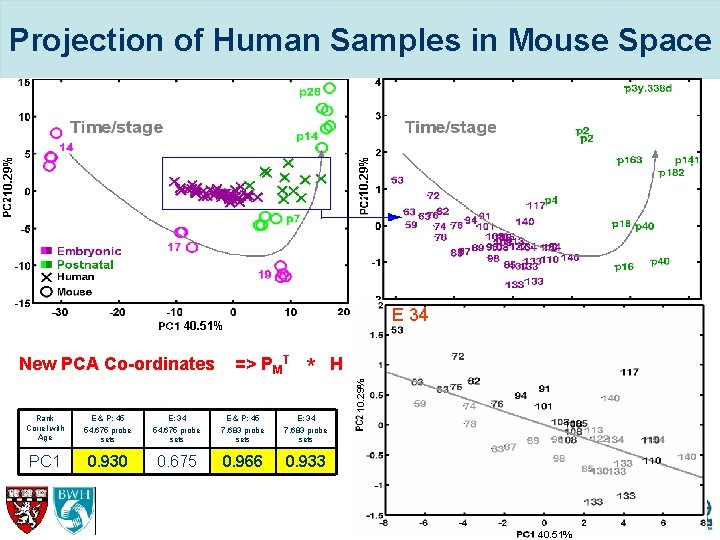
Projection of Human Samples in Mouse Space 10. 29% E & P 45 E 34 40. 51% => PMT * H 10. 29% New PCA Co-ordinates 40. 51% Rank Correl with Age E & P: 45 54, 675 probe sets E: 34 54, 675 probe sets E & P: 45 7, 683 probe sets E: 34 7, 683 probe sets PC 1 0. 930 0. 675 0. 966 0. 933 40. 51%

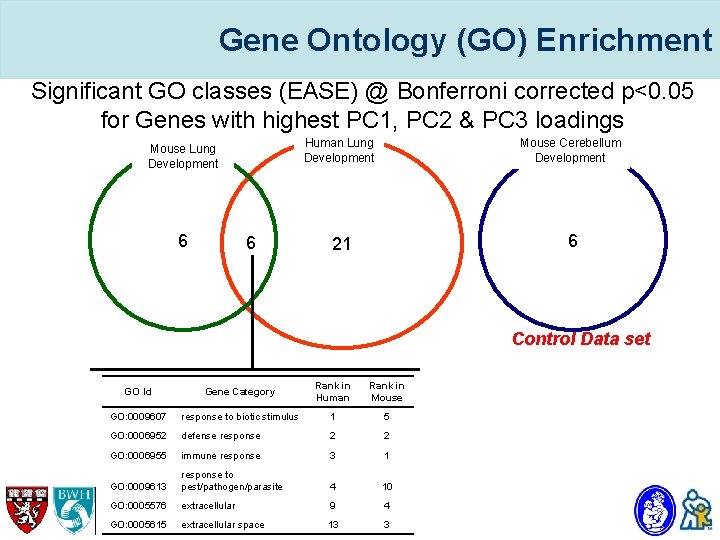
Gene Ontology (GO) Enrichment Significant GO classes (EASE) @ Bonferroni corrected p<0. 05 for Genes with highest PC 1, PC 2 & PC 3 loadings Mouse Lung Development 6 6 Human Lung Development Mouse Cerebellum Development 21 6 Control Data set GO Id Gene Category Rank in Human Rank in Mouse GO: 0009607 response to biotic stimulus 1 5 GO: 0006952 defense response 2 2 GO: 0006955 immune response 3 1 GO: 0009613 response to pest/pathogen/parasite 4 10 GO: 0005576 extracellular 9 4 GO: 0005615 extracellular space 13 3

Conclusions • We observed large-scale concordance between human and murine developing lung transcriptome. • PCA-based change-of-coordinates approach is applicable for general cross-systems multi-scalar model assessment.