Covaris Adaptive Focused Acoustics AFA Optimizing Ch IP













- Slides: 58

Covaris Adaptive Focused Acoustics® (AFA®) Optimizing Ch. IP Sample Preparation for Reproducibility Hamid Khoja Ph. D. Principal Scientist, Covaris Inc. May 30, 2019 www. covaris. com

What is Ch. IP and Epigenetics Proprietary 2

Background Definitions • Chromatin • A complex of DNA, Histones, and other DNA associated proteins which make up the chromosomes • Histones • Highly conserved Proteins which package and order eukaryotic DNA into nucleosomes • ~1. 8 m of DNA in each human cell which is then packaged by histones to about 120 um in chromosomes • Nucleosomes • A histone octamer based spool around which 147 bp of DNA is bound • Genotype • Genetic makeup of a cell/organism • Phenotype • The observable or measurable effect of the genotype in the organism • Chromatin Immunoprecipitation(Ch. IP) is using an antibody to a known protein to immunoprecipitate the DNA that is bound, directly or indirectly to that protein. Proprietary 3

Ch. IP and Epigenetics Research go hand in hand • Epigenetics can be defined in its simples form as a study of extra-genomic pathways to phenotype differences in organisms. • All human cells contains the same DNA, yet vast number of different cell types exist which look and function quite differently from one another. • Identical twins with distinguishable features is an example of environmentally induced epigenetics • Achieved by proteins directly or indirectly binding to and modifying DNA or DNA associated proteins, and affecting the level of RNA expression (transcription), as well as subsequent translation( protein synthesis) and post translational modification of proteins. Proprietary 4

Ch. IP History Proprietary 5

Ch. IP History 1985 – UV-crosslinked chromatin used to study distribution of RNA polymerase II on fruit fly heat shock genes John Tis and David Gilmour, Molecular and Cellular Biology 1988 – Formaldehyde cross linked chromatin used for the first time to study histone H 4 distribution heat shock genes Alexander Varshavsky, Cell 1997 – Analysis of chromatin structure by in vivo formaldehyde crosslinking Valerio Orlando, Nature Methods The sample preparation has not changed much for the past 30 years!! Proprietary 6

Historically used Shearing Tools Historically shearing has been achieved using: • High energy probe sonicators • High energy bath sonicators • Enzymatic digestion Proprietary 7

8 Chromatin Shearing Methods • Enzymatic • Fragmenting bias • Low resolution • Fixation-caused steric hindrance • Bath sonication (including Cup horn) • • High energy Heat control issues Reproducibility issues Transducer degrades over time • Probe sonication • • Vibrating hot metal rod in contact with limited sample volume Thermal control Cross contamination issues Probe degrades over time Proprietary 8

Continued utility of these Technologies for Ch. IP? • Antiquated technologies that have reached their absolute potential for chromatin shearing • Technologies NOT developed with reproducible shearing in mind • Merely adaptation of old technologies which have been around for decades • Lack accuracy and precision • Lack of control over process • Limited automation and throughput capabilities Proprietary 9

Outdated Chromatin Shearing Protocols Most published protocols were developed using high energy, uncontrolled sonicators, which: • Required - High cell numbers • Compensation for low chromatin yield • Required - Longer formaldehyde fixation • Compensate for the destructive nature of high energy shearing • Exposed sample to uncontrolled temperature fluctuations • Consequence of high energy uncontrolled processing …Limited mostly to abundant DNA binding proteins Proprietary 10

Ch. IP Workflow and Success Metrics Proprietary 11

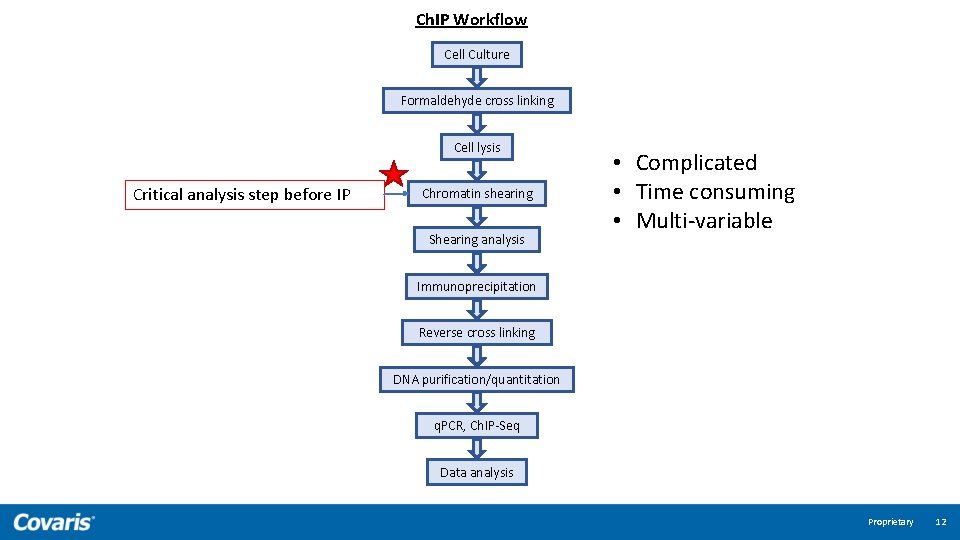
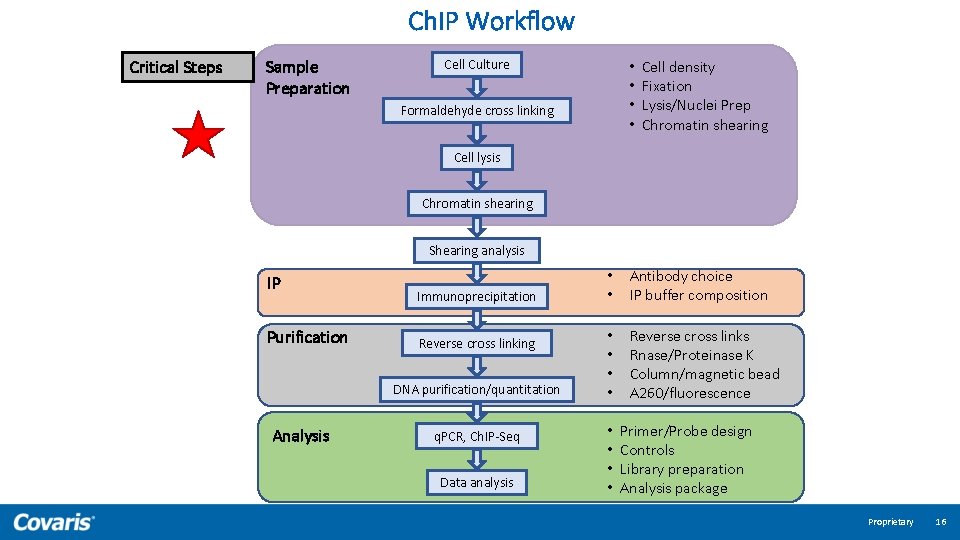
Ch. IP Workflow Cell Culture Formaldehyde cross linking Cell lysis Critical analysis step before IP Chromatin shearing Shearing analysis • Complicated • Time consuming • Multi-variable Immunoprecipitation Reverse cross linking DNA purification/quantitation q. PCR, Ch. IP-Seq Data analysis Proprietary 12

Characteristics of Reproducible Chromatin Shearing • Precise • Energy control to obtain desired fragment size • Accurate • Thermal control maintain epitope integrity • Controlled • Optimized instrument, protocol, and reagents system Proprietary 13

Importance of Reproducible Chromatin Shearing • • Generate an unbiased representation of the entire original chromatin Conserve Ch. IP epitopes Treated and untreated samples processed identically Generate fragment sizes usable in the preparation of libraries for all NGS systems Proprietary 14

Critical Steps of Ch. IP Sample Preparation Proprietary 15

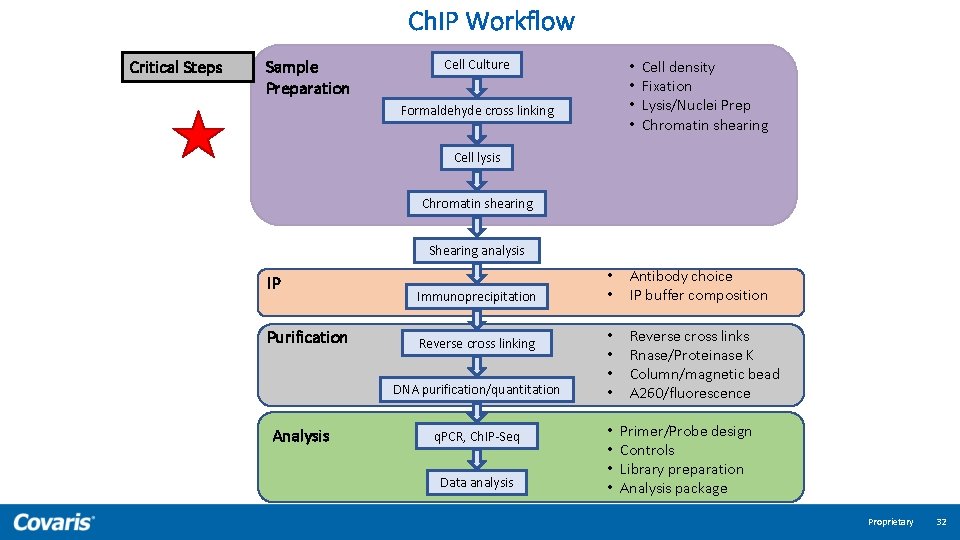
Ch. IP Workflow Critical Steps Sample Preparation Cell Culture • • Formaldehyde cross linking Cell density Fixation Lysis/Nuclei Prep Chromatin shearing Cell lysis Chromatin shearing Shearing analysis IP Purification Immunoprecipitation Reverse cross linking DNA purification/quantitation Analysis q. PCR, Ch. IP-Seq Data analysis • • Antibody choice IP buffer composition • • Reverse cross links Rnase/Proteinase K Column/magnetic bead A 260/fluorescence • • Primer/Probe design Controls Library preparation Analysis package Proprietary 16

1 7 Three Critical Steps of Ch. IP Sample Preparation 1. Formaldehyde Fixation • Covalently link and preserve protein-DNA interaction for immunoprecipitation 2. Lysis • Prepare nuclei and lyse for chromatin release 3. Shearing • Fragmenting chromatin to desired size range for immunoprecipitation Proprietary 17

1 8 Formaldehyde Fixation Fix cells sufficiently to maintain protein-DNA interaction during shearing, but “loose” enough to allow reversing of cross links and isolate DNA after IP . . . A great majority of people over-fix their samples. . Because they never optimize the fixation step. Proprietary 18

1 9 Formaldehyde Fixation Optimization • Choosing the correct formaldehyde formulations • Powdered paraformaldehyde • Dangerous and cumbersome preparation • Generates complex mixture of formaldehyde polymers (HCOH)n. . . • Formalin polymers have varying fixation rates • Concentrated 37% formaldehyde • Stabilized with 15% methanol to reduce polymerization rate • Methanol evaporation over time allow formaldehyde polymerization • Methanol increases cell permeability rendering fixation more potent • Methanol-free 16% formaldehyde • Contain formaldehyde monomers only • Single use ampoules • Provides the most precise and accurate results Proprietary 19

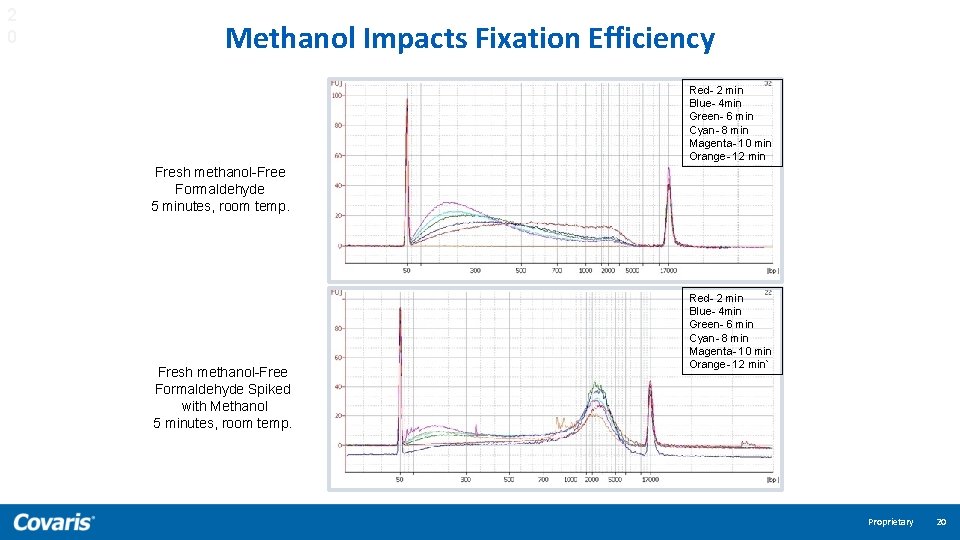
2 0 Methanol Impacts Fixation Efficiency Red- 2 min Blue- 4 min Green- 6 min Cyan- 8 min Magenta- 10 min Orange- 12 min Fresh methanol-Free Formaldehyde 5 minutes, room temp. Fresh methanol-Free Formaldehyde Spiked with Methanol 5 minutes, room temp. Red- 2 min Blue- 4 min Green- 6 min Cyan- 8 min Magenta- 10 min Orange- 12 min` Proprietary 20

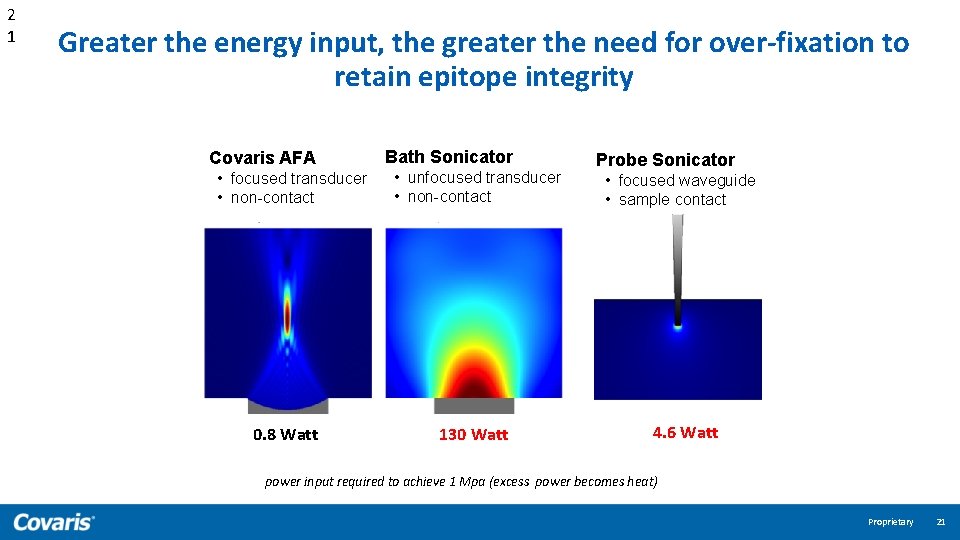
2 1 Greater the energy input, the greater the need for over-fixation to retain epitope integrity Covaris AFA • focused transducer • non-contact 0. 8 Watt Bath Sonicator • unfocused transducer • non-contact 130 Watt Probe Sonicator • focused waveguide • sample contact 4. 6 Watt power input required to achieve 1 Mpa (excess power becomes heat) Proprietary 21

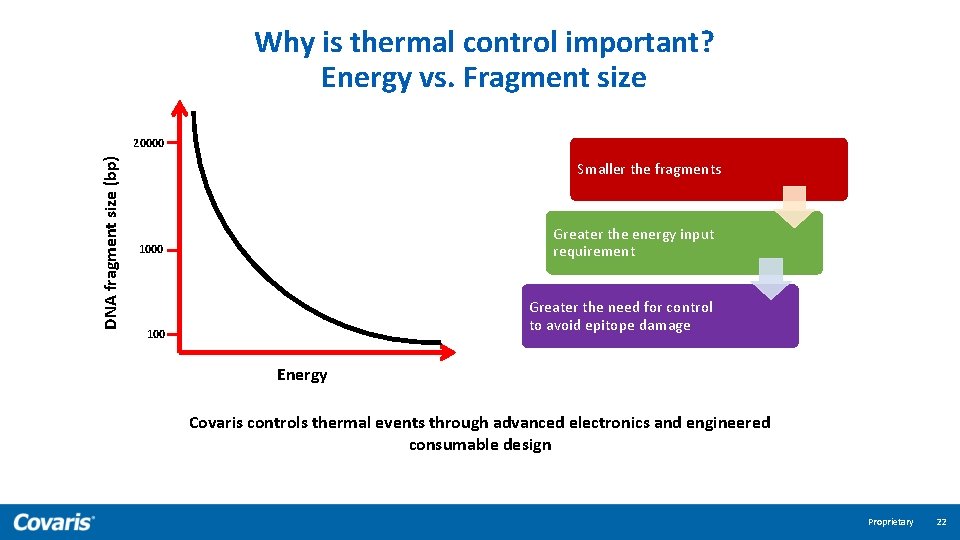
Why is thermal control important? Energy vs. Fragment size DNA fragment size (bp) 20000 Smaller the fragments Greater the energy input requirement 1000 Greater the need for control to avoid epitope damage 100 Energy Covaris controls thermal events through advanced electronics and engineered consumable design Proprietary 22

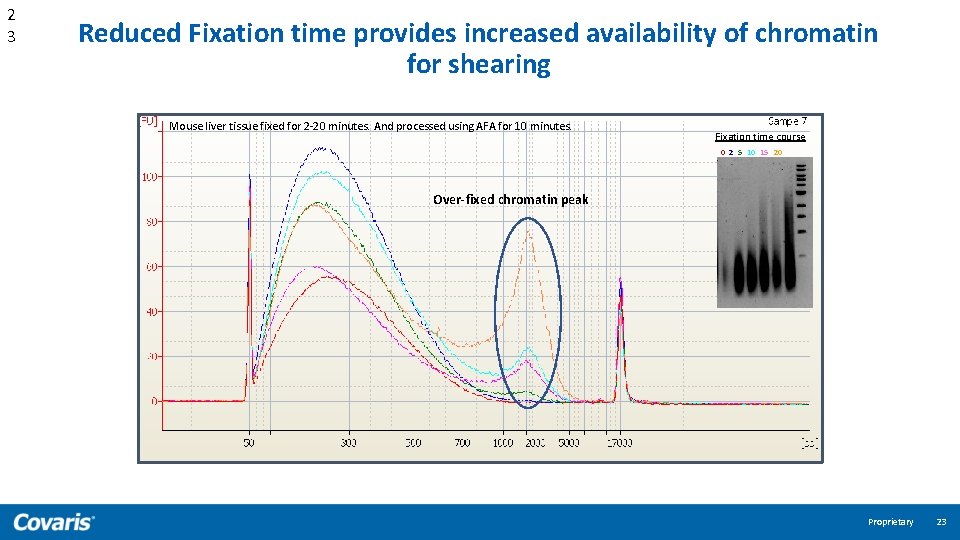
2 3 Reduced Fixation time provides increased availability of chromatin for shearing Mouse liver tissue fixed for 2 -20 minutes. And processed using AFA for 10 minutes. Fixation time course 0 2 5 10 15 20 Over-fixed chromatin peak Proprietary 23

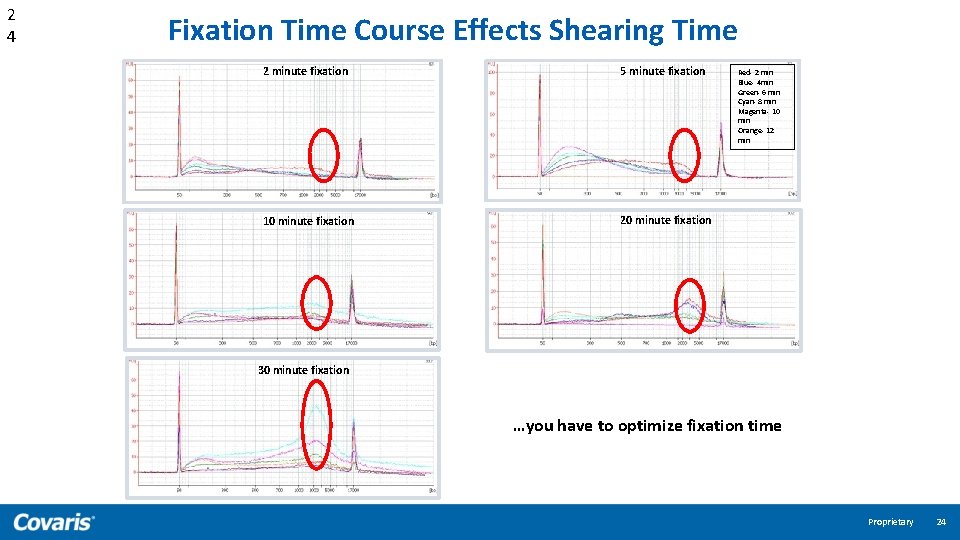
2 4 Fixation Time Course Effects Shearing Time 2 minute fixation 5 minute fixation 10 minute fixation 20 minute fixation Red- 2 min Blue- 4 min Green- 6 min Cyan- 8 min Magenta- 10 min Orange- 12 min 30 minute fixation …you have to optimize fixation time Proprietary 24

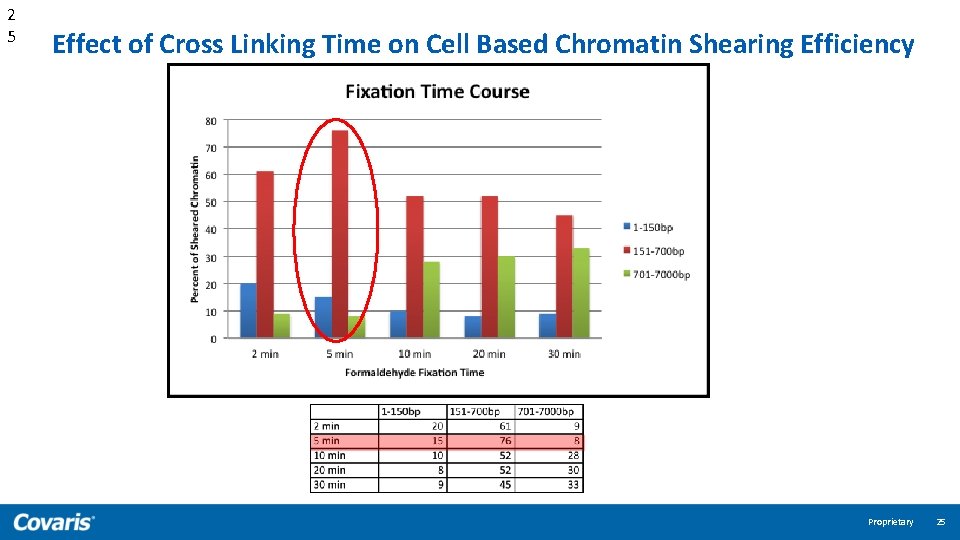
2 5 Effect of Cross Linking Time on Cell Based Chromatin Shearing Efficiency Proprietary 25

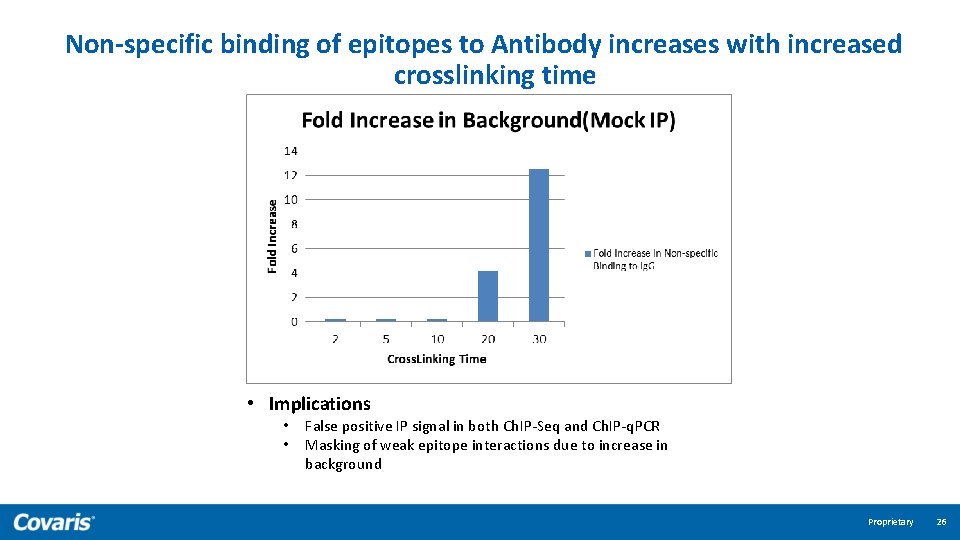
Non-specific binding of epitopes to Antibody increases with increased crosslinking time • Implications • • False positive IP signal in both Ch. IP-Seq and Ch. IP-q. PCR Masking of weak epitope interactions due to increase in background Proprietary 26

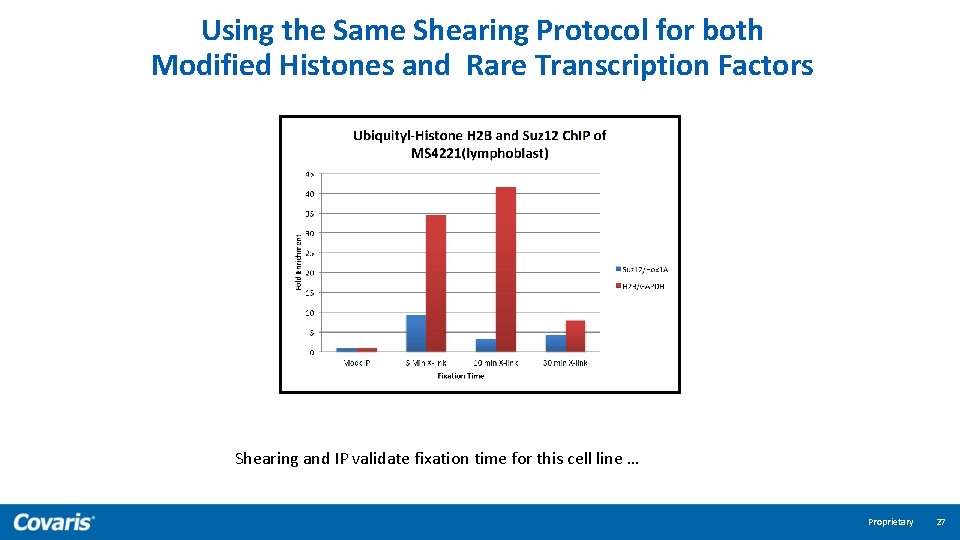
Using the Same Shearing Protocol for both Modified Histones and Rare Transcription Factors Shearing and IP validate fixation time for this cell line … Proprietary 27

2 8 Fixation Optimization • Fixation time Optimization • Cell line dependent • All Cells do not fix the same way. . • Optimal fixation time has to be determined empirically • Epitope dependent • All proteins do not fix the same way to DNA. . • Over-fixation can effect antibody binding • Formaldehyde concentration • 1% concentration is now the standard for chromatin fixation • Lower concentrations used when processing <100 k cells Proprietary 28

Cell Lysis Methodologies used for Ch. IP • One step lysis/shearing • Require high detergent concentrations • Works only on certain cell lines • High energy shearing required • Osmotic lysis • Works well with large cells such as He. La and Hep. G 2 but not with small cells such as Jurkat, 293, and primary cells • Mild multi-detergent lysis for nuclei preparation • The most efficient method of nuclei preparation • Works universally with all mammalian cell types • Allows use of low detergent concentration use in shearing buffer • Sheared chromatin usable with any kit and homebrew IP methods Proprietary 29

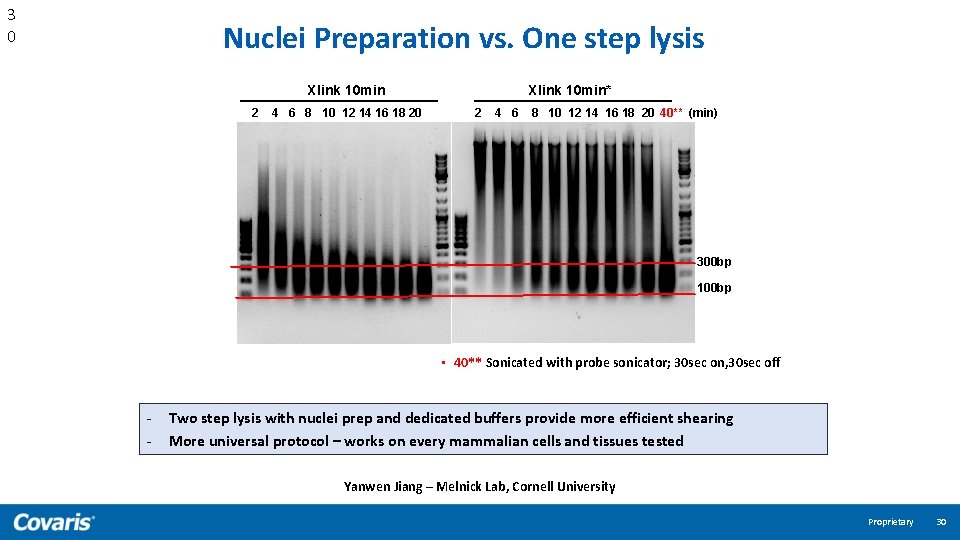
3 0 Nuclei Preparation vs. One step lysis Xlink 10 min 2 4 6 8 10 12 14 16 18 20 Xlink 10 min* 2 4 6 8 10 12 14 16 18 20 40** (min) 300 bp 100 bp • 40** Sonicated with probe sonicator; 30 sec on, 30 sec off - Two step lysis with nuclei prep and dedicated buffers provide more efficient shearing More universal protocol – works on every mammalian cells and tissues tested Yanwen Jiang – Melnick Lab, Cornell University Proprietary 30

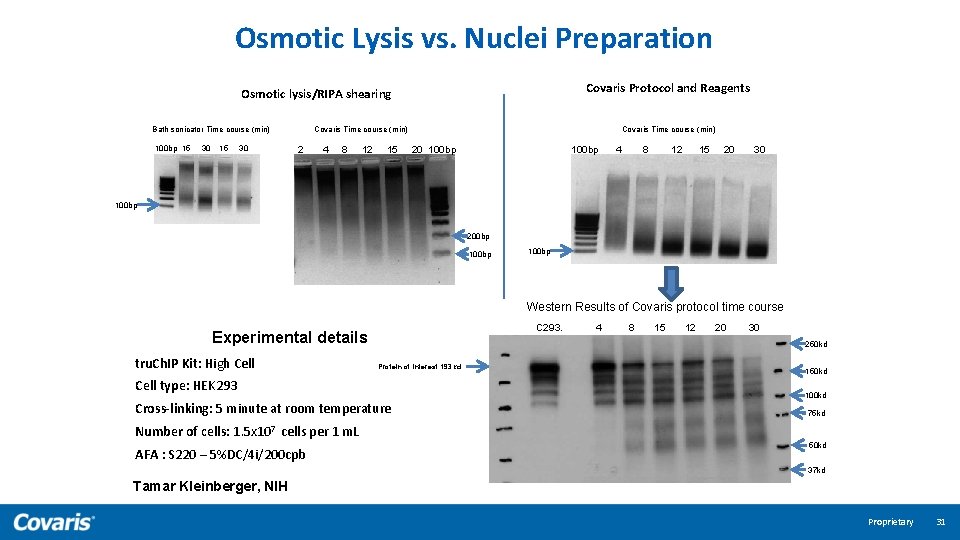
Osmotic Lysis vs. Nuclei Preparation Covaris Protocol and Reagents Osmotic lysis/RIPA shearing Covaris Time course (min) Bath sonicator Time course (min) 100 bp 15 30 2 4 8 12 15 Covaris Time course (min) 100 bp 20 100 bp 4 8 12 15 20 30 100 bp 200 bp 100 bp Western Results of Covaris protocol time course C 293. Experimental details tru. Ch. IP Kit: High Cell 4 8 15 12 20 30 250 kd Protein of interest 193 kd Cell type: HEK 293 Cross-linking: 5 minute at room temperature 150 kd 100 kd 75 kd Number of cells: 1. 5 x 107 cells per 1 m. L AFA : S 220 – 5%DC/4 i/200 cpb 50 kd 37 kd Tamar Kleinberger, NIH Proprietary 31

Ch. IP Workflow Critical Steps Sample Preparation Cell Culture • • Formaldehyde cross linking Cell density Fixation Lysis/Nuclei Prep Chromatin shearing Cell lysis Chromatin shearing Shearing analysis IP Purification Immunoprecipitation Reverse cross linking DNA purification/quantitation Analysis q. PCR, Ch. IP-Seq Data analysis • • Antibody choice IP buffer composition • • Reverse cross links Rnase/Proteinase K Column/magnetic bead A 260/fluorescence • • Primer/Probe design Controls Library preparation Analysis package Proprietary 32

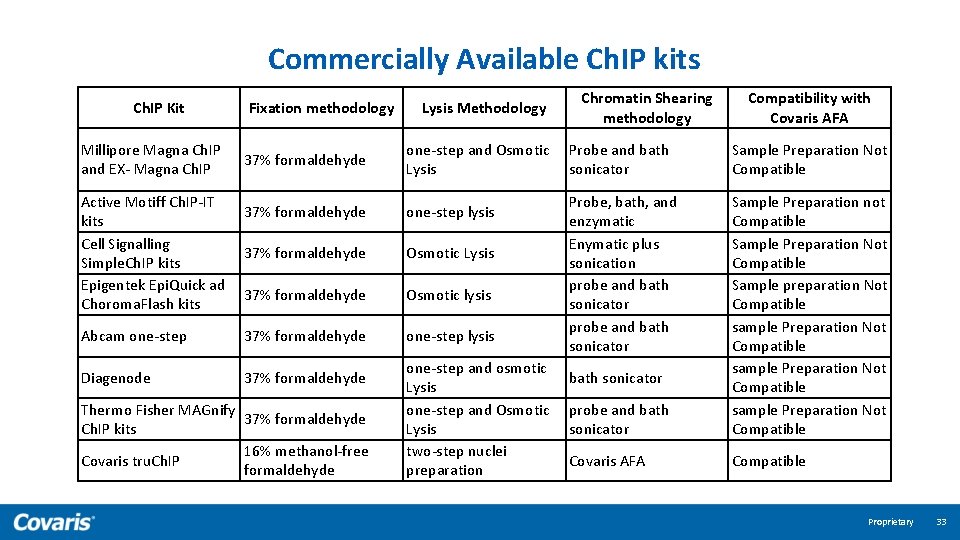
Commercially Available Ch. IP kits Ch. IP Kit Millipore Magna Ch. IP and EX- Magna Ch. IP Fixation methodology 37% formaldehyde Active Motiff Ch. IP-IT 37% formaldehyde kits Cell Signalling 37% formaldehyde Simple. Ch. IP kits Epigentek Epi. Quick ad 37% formaldehyde Choroma. Flash kits Abcam one-step 37% formaldehyde Diagenode 37% formaldehyde Thermo Fisher MAGnify 37% formaldehyde Ch. IP kits 16% methanol-free Covaris tru. Ch. IP formaldehyde Lysis Methodology Chromatin Shearing methodology one-step and Osmotic Probe and bath Lysis sonicator one-step lysis Osmotic Lysis Osmotic lysis one-step and osmotic Lysis one-step and Osmotic Lysis two-step nuclei preparation Probe, bath, and enzymatic Enymatic plus sonication probe and bath sonicator Compatibility with Covaris AFA Sample Preparation Not Compatible probe and bath sonicator Sample Preparation not Compatible Sample Preparation Not Compatible Sample preparation Not Compatible sample Preparation Not Compatible Covaris AFA Compatible bath sonicator Proprietary 33

Critical Step in Ch. IP: Chromatin shearing Good Shearing Bad Shearing Proceed to IP Discard, change a variable and repeat Proprietary 34

Covaris tru. Ch. IP: A Different Approach to Chromatin Shearing • Lower cell number requirement • Reduced cell culturing • Controlled shearing size range • Usable for all NGS platform library preparation • Isothermal processing • Maintaining epitope integrity • Low detergent requirement • Suitable for all IP protocols without sample dilution • Lower fixation time requirement • Availability of more chromatin for IP • Universal protocol guaranteed to work with all mammalian cells …which enables highly reproducible shearing Proprietary 35

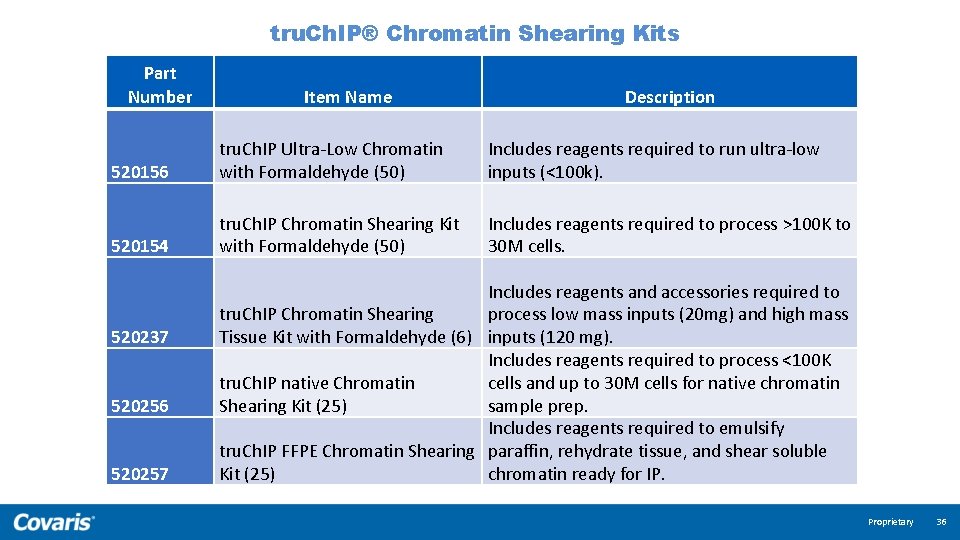
tru. Ch. IP® Chromatin Shearing Kits Part Number Item Name Description 520156 tru. Ch. IP Ultra-Low Chromatin with Formaldehyde (50) Includes reagents required to run ultra-low inputs (<100 k). 520154 tru. Ch. IP Chromatin Shearing Kit with Formaldehyde (50) Includes reagents required to process >100 K to 30 M cells. 520237 520256 520257 Includes reagents and accessories required to process low mass inputs (20 mg) and high mass tru. Ch. IP Chromatin Shearing Tissue Kit with Formaldehyde (6) inputs (120 mg). Includes reagents required to process <100 K cells and up to 30 M cells for native chromatin tru. Ch. IP native Chromatin sample prep. Shearing Kit (25) Includes reagents required to emulsify tru. Ch. IP FFPE Chromatin Shearing paraffin, rehydrate tissue, and shear soluble chromatin ready for IP. Kit (25) Proprietary 36

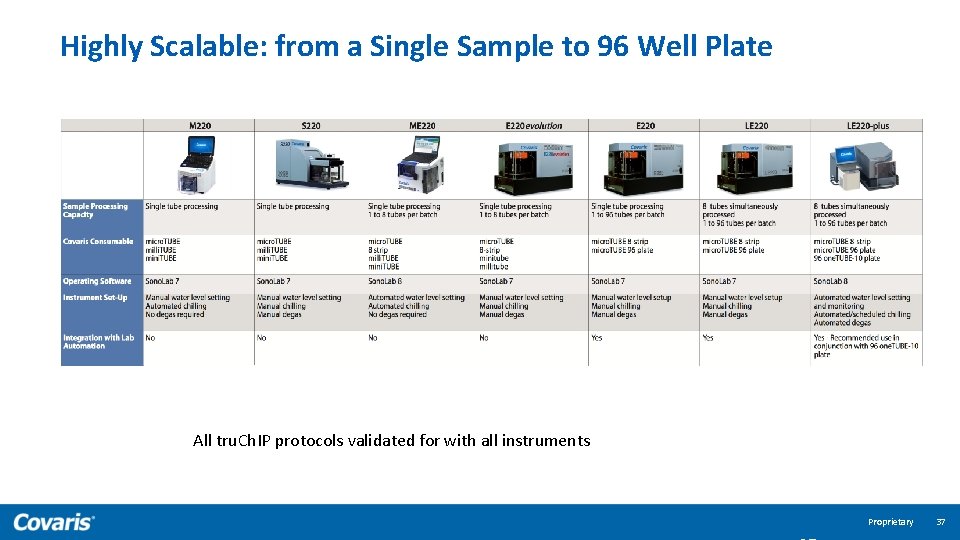
Highly Scalable: from a Single Sample to 96 Well Plate Compatible for use with all Covaris Instruments All tru. Ch. IP protocols validated for with all instruments Proprietary 37

Fields of Research and Potential Customers Proprietary 38

Fields of Research that fall into the sphere of Epigenetics • Cancer Research • Inherited • • Sporadic ( non-inherited) caner • • Cell differentiation/programming • External stimuli affecting gene regulation Viral infection • • Elucidation of the intricate cascade of replication and error/spontaneous mutation repair Cell signaling/signal transduction • • Control of gene transcription DNA replication/repair/modification • • Identification of gene products which determine cell differentiation and maintain a differentiated state Gene Regulation • • Epigenetic changes initiating somatic changes in a cell Stem cell research • • Genetic mutations initiate cancer but epigenetic changes promote its progression Hijacking/bypassing of a cells replication/translation/transcription machinery to synthesize viral particles Small molecule research • Up and down regulation of genes using synthetic compounds Proprietary 39

DNA-protein interaction assays that use NGS and can benefit from AFA and tru. Ch. IP • Ch. IP-seq • FAIRE-seq ( Formaldehyde Assisted Isolation of Regulatory Elements) • Used to determine only the non-protein bound regions of the chromatin • Ch. IAPET-Seq( Chromatin Interaction Analysis by Paired End Tag) • Long range interaction of chromatin with specific proteins of interest • HI-C/3 C-seq (Chromatin Conformation Capture) • Determining long range chromatin interactions • 4 C-seq (Circular Chromatin Conformation Capture) • Unbiased detection of all genomic regions that interact with a particular region of interest • 5 C-seq ( Chromatin Conformation Capture Carbon Copy) • Concurrent determination of interactions between multiple sequences. A higher throughput version of C 4 Proprietary 40

RNA-protein interaction assays that use NGS and can benefit from AFA and tru. Ch. IP • Ch. IRP-seq ( Chromatin Isolation by RNA Purification) • Determining where non coding RNAs(nc. RNA) and their proteins are bound to the genome • RIP-seq ( RNA Immunoprecipitation) • Mapping the sites where proteins are bound to RNA in RNA-protein complexes • CLIP-seq (Cross Linking and Immunoprecipitation) • Mapping of protein-RNA binding sites • ICLIP-seq (Individual nucleotide resolution Cross Linking and Immunoprecipitation) • High resolution mapping of protein-RNA binding sites • CLASH-seq ( Crosslinking, Ligation, and Sequencing of Hybrids) • Mapping of RNA-RNA interactions with a specific protein Proprietary 41

DNA methylation applications that use NGS and can benefit from AFA • MEDIP-seq (Methylated DNA Immunoprecipitation) • Mapping of 5 m. C modification sites on the genome • BS-seq ( Bi. Sulfite) • Mapping of methylated cytosines in the genome Proprietary 42

Publications from Key Opinion Leaders Proprietary 43

Supported Chromatin-based NGS Applications Ch. IP-Seq Ch. IA-PET Hi-C Adaptive Focused AcousticsⓇ (AFAⓇ) Methyl-Seq Ch. IRP FAIRE-Seq Proprietary 44

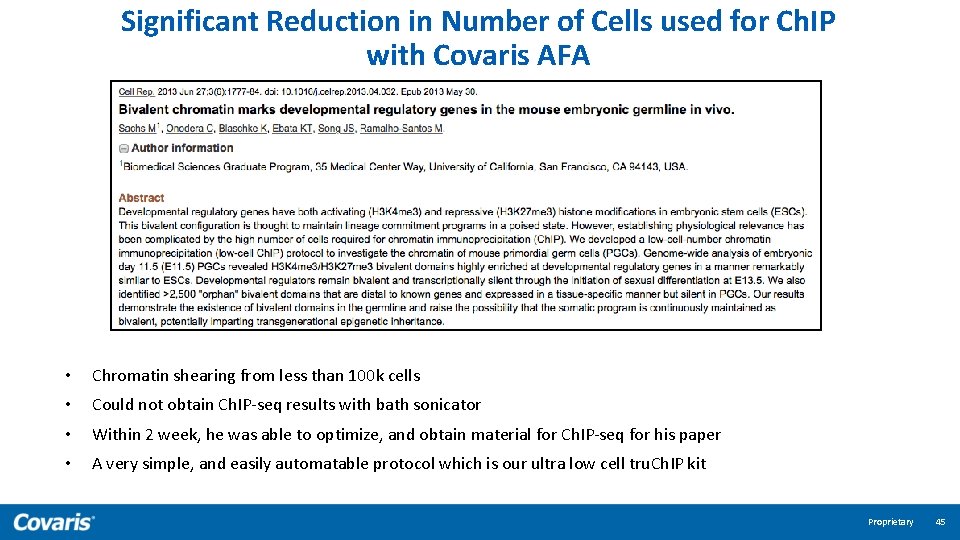
Significant Reduction in Number of Cells used for Ch. IP with Covaris AFA • Chromatin shearing from less than 100 k cells • Could not obtain Ch. IP-seq results with bath sonicator • Within 2 week, he was able to optimize, and obtain material for Ch. IP-seq for his paper • A very simple, and easily automatable protocol which is our ultra low cell tru. Ch. IP kit Proprietary 45

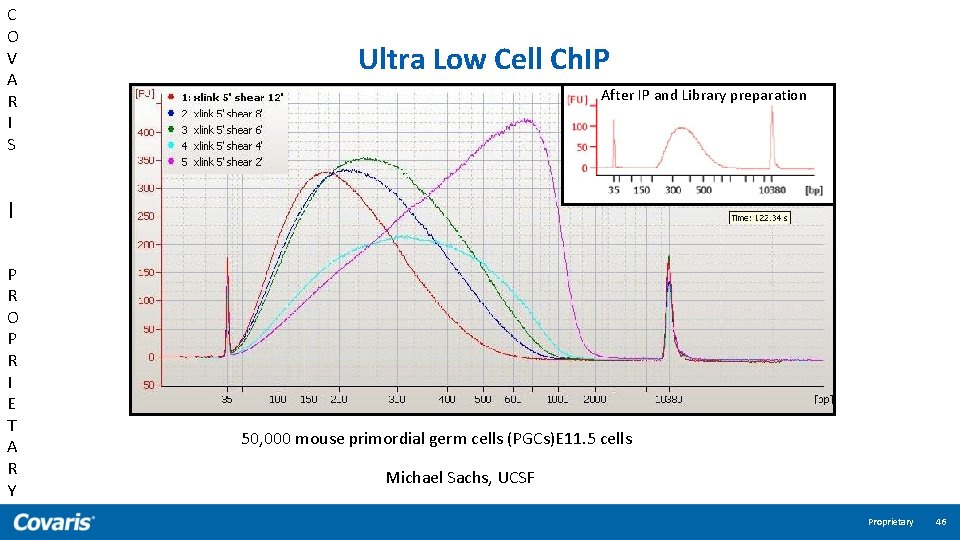
C O V A R I S | P R O P R I E T A R Y Ultra Low Cell Ch. IP After IP and Library preparation 50, 000 mouse primordial germ cells (PGCs)E 11. 5 cells Michael Sachs, UCSF Proprietary 46

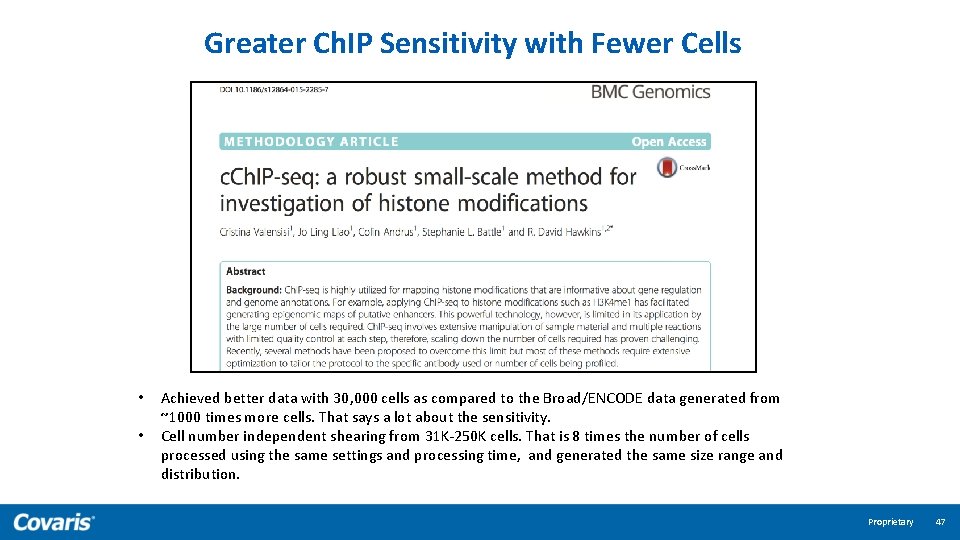
Greater Ch. IP Sensitivity with Fewer Cells • • Achieved better data with 30, 000 cells as compared to the Broad/ENCODE data generated from ~1000 times more cells. That says a lot about the sensitivity. Cell number independent shearing from 31 K-250 K cells. That is 8 times the number of cells processed using the same settings and processing time, and generated the same size range and distribution. Proprietary 47

Active Cell Lysis for Higher Chromatin Yield • Nuclei preparation carried out with Covaris tru. Ch. IP lysis buffer • Active cell lysis using Covaris AFA to increase nuclei yield • Shearing carried out in tru. Ch. IP SDS shearing buffer Proprietary 48

DNA Methylation Analysis Proprietary 49

Ch. IRP: nc. RNA interactions with chromatin Proprietary 50

Hi. C: Key Publications in 3 D Chromatin Organization Proprietary 51

Capture-C : 3 D Chromatin Organization Proprietary 52

Ch. IA-PET: Investigation of Proteins Involved in 3 D chromatin Organization Proprietary 53

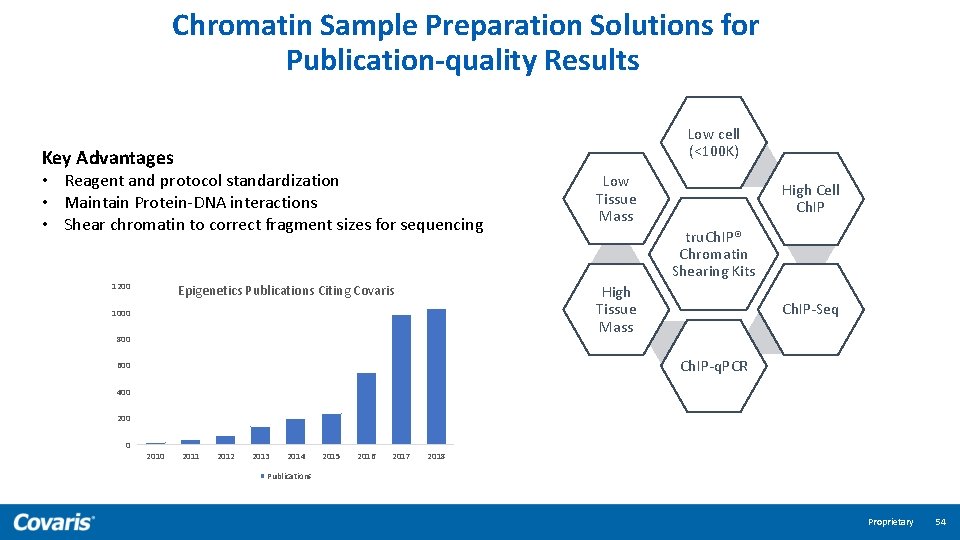
Chromatin Sample Preparation Solutions for Publication-quality Results Low cell (<100 K) Key Advantages • Reagent and protocol standardization • Maintain Protein-DNA interactions • Shear chromatin to correct fragment sizes for sequencing 1200 Epigenetics Publications Citing Covaris Low Tissue Mass High Cell Ch. IP tru. Ch. IP® Chromatin Shearing Kits High Tissue Mass 1000 800 Ch. IP-Seq Ch. IP-q. PCR 600 400 200 0 2011 2012 2013 2014 2015 2016 2017 2018 Publications Proprietary 54

Example Publications on Drosophila Ch. IP Proprietary 55

Example Publications on Yeast and Plant Ch. IP Proprietary 56

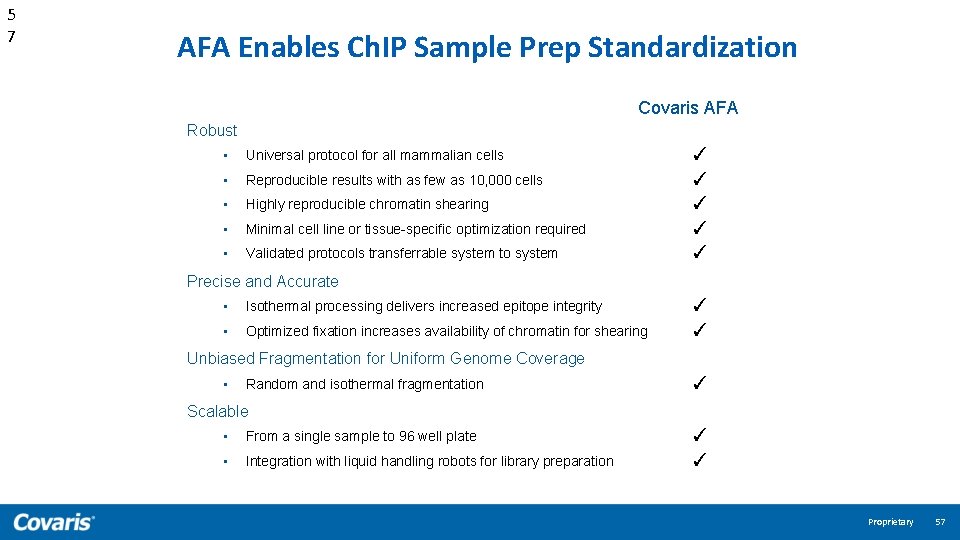
5 7 AFA Enables Ch. IP Sample Prep Standardization Covaris AFA Robust • Universal protocol for all mammalian cells • Reproducible results with as few as 10, 000 cells • Highly reproducible chromatin shearing • Minimal cell line or tissue-specific optimization required • Validated protocols transferrable system to system ✓ ✓ ✓ Precise and Accurate • Isothermal processing delivers increased epitope integrity • Optimized fixation increases availability of chromatin for shearing ✓ ✓ Unbiased Fragmentation for Uniform Genome Coverage • Random and isothermal fragmentation ✓ Scalable • From a single sample to 96 well plate • Integration with liquid handling robots for library preparation ✓ ✓ Proprietary 57

Thank you • Hamid Khoja, Principal Scientist hkhoja@covarisinc. com • Sales and orders Support: Customer. Service@covarisinc. com • Protocol, method and application support: Application. Support@covarisinc. com • instruments and consumables: Tech. Support@covarisinc. com Proprietary 58