COT 6930 HPC and Bioinformatics Protein Structure Prediction
COT 6930 HPC and Bioinformatics Protein Structure Prediction Xingquan Zhu Dept. of Computer Science and Engineering A. Y. Zhang, University of Kansas
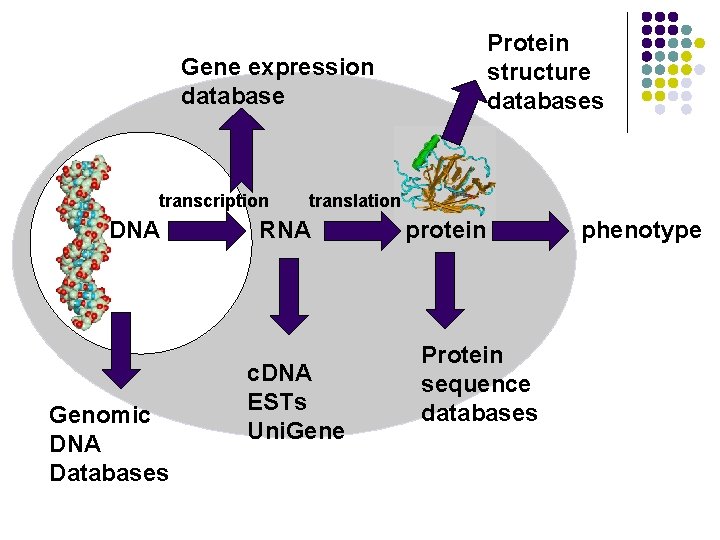
Protein structure databases Gene expression database transcription DNA Genomic DNA Databases translation RNA c. DNA ESTs Uni. Gene protein Protein sequence databases phenotype
Outline l Protein Structure l l l Why structure How to predict protein structure l Experimental methods l Computational methods (predictive methods) Protein Structure Prediction l Secondary structure prediction (2 D) l l Machine learning methods for protein secondary structure prediction Tertiary structure prediction (3 D) l Ab initio l Homology modeling

Proteins l l Proteins play a crucial role in virtually all biological processes with a broad range of functions. The activity of an enzyme or the function of a protein is governed by the three-dimensional structure
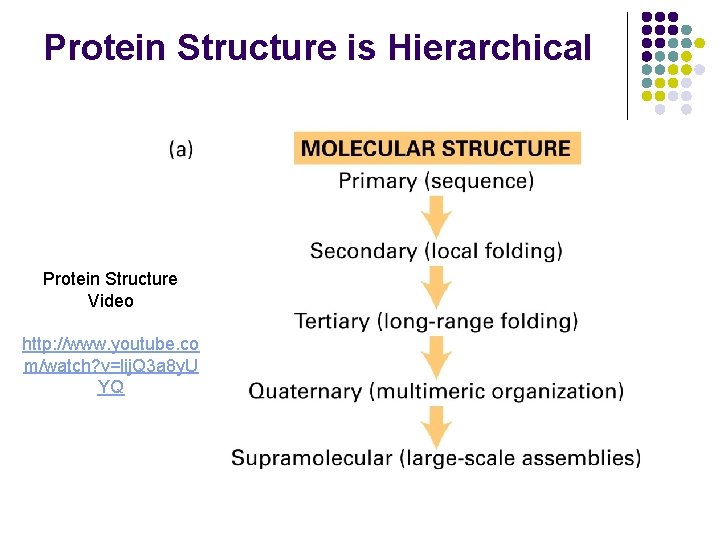
Protein Structure is Hierarchical Protein Structure Video http: //www. youtube. co m/watch? v=lij. Q 3 a 8 y. U YQ
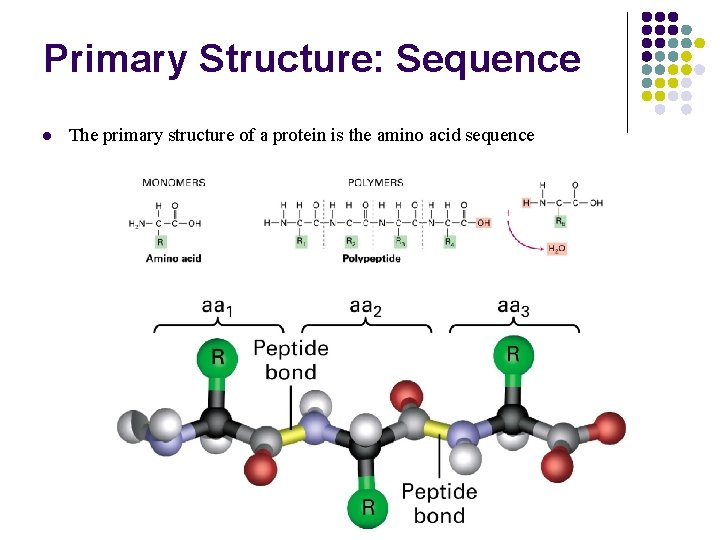
Primary Structure: Sequence l The primary structure of a protein is the amino acid sequence

Protein Structure Prediction Problem Protein structure prediction l l Predict protein 3 D structure from (amino acid) sequence One step closer to useful biological knowledge Sequence → secondary structure → 3 D structure → function

Outline l Protein Structure l l l Why structure How to Predict Protein Structure l Experimental methods l Computational methods (predictive methods) Protein Structure Prediction l Secondary structure prediction (2 D) l l Machine learning methods for Protein Secondary Structure Prediction Tertiary structure prediction (3 D) l Ab initio l Homology modeling

Why Predict Structure? Structure is more conserved than sequence Structure determines function Goals: 1. Predict structure from sequence 2. Predict function based on structure 3. Predict function based on sequence Molecular function

Why predict structure: Structure is more conserved than sequence 28% sequence identity

Why predict structure: Can Label Proteins by Dominant Structure l SCOP: Structural Classification Of Proteins
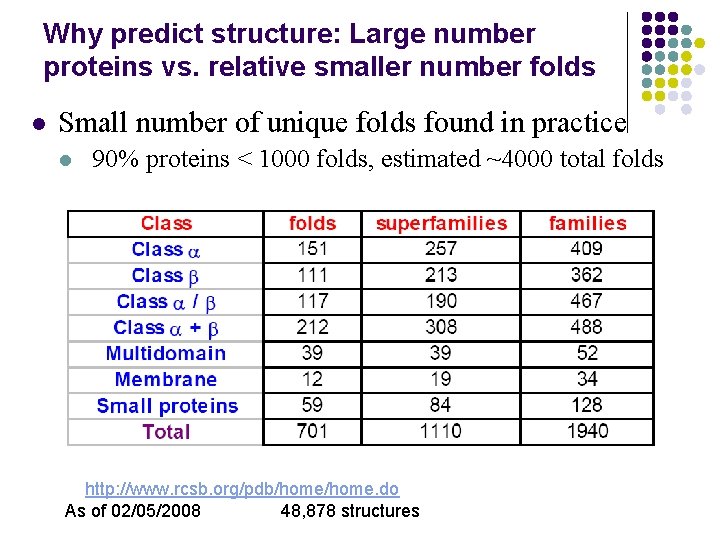
Why predict structure: Large number proteins vs. relative smaller number folds l Small number of unique folds found in practice l 90% proteins < 1000 folds, estimated ~4000 total folds http: //www. rcsb. org/pdb/home. do As of 02/05/2008 48, 878 structures
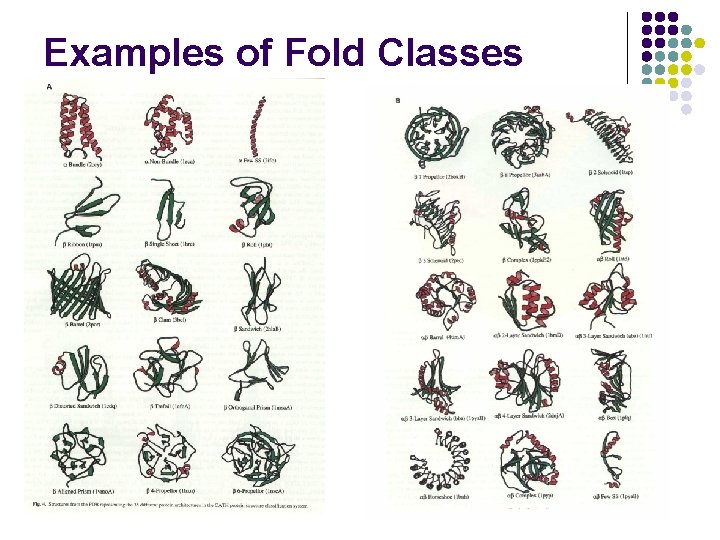
Examples of Fold Classes
How to Predict Protein Structure l A related biological question: what are the factors that determine a structure? l l l Energy Kinematics How can we determine structure? l Experimental methods l X-ray crystallography or NMR (Nuclear magnetic resonance) spectrometry § l limitation: protein size, require crystallized proteins Computational methods (predictive methods) l 2 -D structure (secondary structure) l 3 -D structure (tertiary structure)
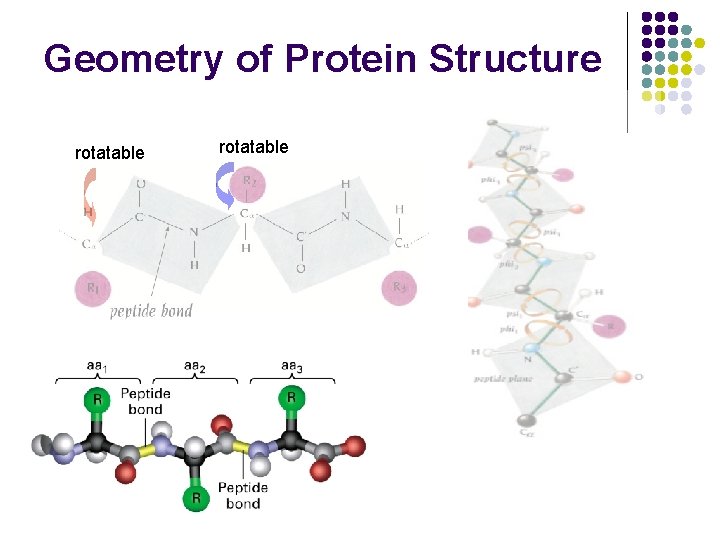
Geometry of Protein Structure rotatable

Inter-atomic Forces l Covalent bond l l l (short range, very strong) Covalent bond between sulfhydryl (sulfur + hydrogen) groups Hydrophobic / hydrophillic interaction (weak) l l (short range, strong) Binds two polar groups (hydrogen + electronegative atom) Disulfide bond / bridge l l Binds atoms into molecules / macromolecules Hydrogen bond l (short range, very strong) Hydrogen bonding w/ H 2 O in solution Van der Waal’s interaction l Nonspecific electrostatic attractive force (very weak)
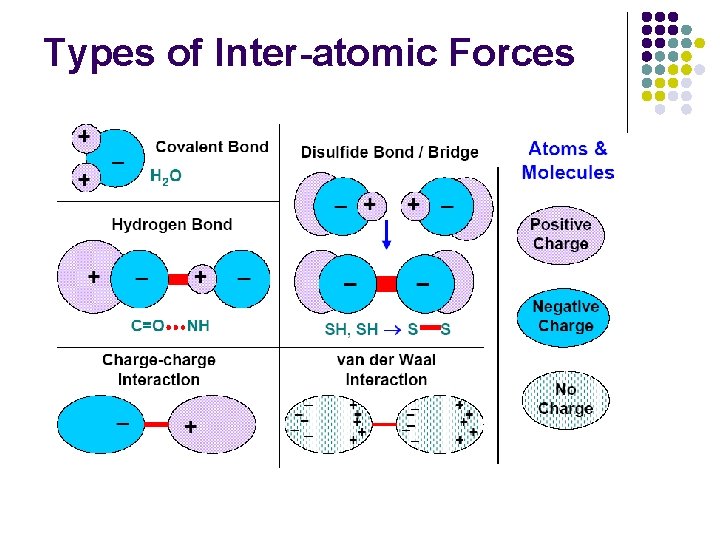
Types of Inter-atomic Forces
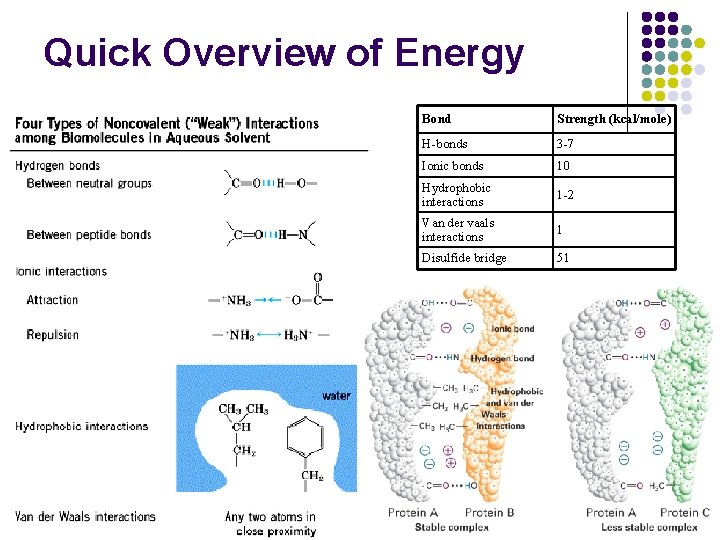
Quick Overview of Energy Bond Strength (kcal/mole) H-bonds 3 -7 Ionic bonds 10 Hydrophobic interactions 1 -2 Van der vaals interactions 1 Disulfide bridge 51

Protein Folding Animation l l http: //www. youtube. com/watch? v=fv. BO 3 Tq. J 6 FE http: //www. youtube. com/watch? v=sw. Ec_s. UVz 5 I

Two Related Problems in Structure Prediction l l Directly predicting protein structure from the amino acid sequence has proved elusive Two sub-problems l l Secondary Structure Prediction Tertiary Structure Prediction
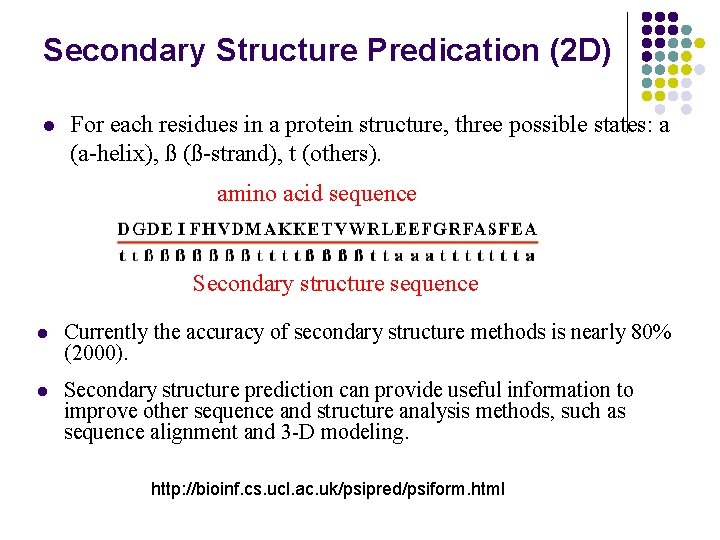
Secondary Structure Predication (2 D) l For each residues in a protein structure, three possible states: a (a-helix), ß (ß-strand), t (others). amino acid sequence Secondary structure sequence l Currently the accuracy of secondary structure methods is nearly 80% (2000). l Secondary structure prediction can provide useful information to improve other sequence and structure analysis methods, such as sequence alignment and 3 -D modeling. http: //bioinf. cs. ucl. ac. uk/psipred/psiform. html

Outline l Protein Structure l l l Why structure How to Predict Protein Structure l Experimental methods l Computational methods (predictive methods) Protein Structure Prediction l Secondary structure prediction (2 D) l l Machine learning methods for Protein Secondary Structure Prediction Tertiary structure prediction (3 D) l Ab initio l Homology modeling
PSSP: Protein Secondary Structure Prediction l Three Generations • • • Based on statistical information of single amino acids Based on local amino acid interaction (segments). Typically a segment containes 11 -21 aminoacids Based on evolutionary information of the homology sequences
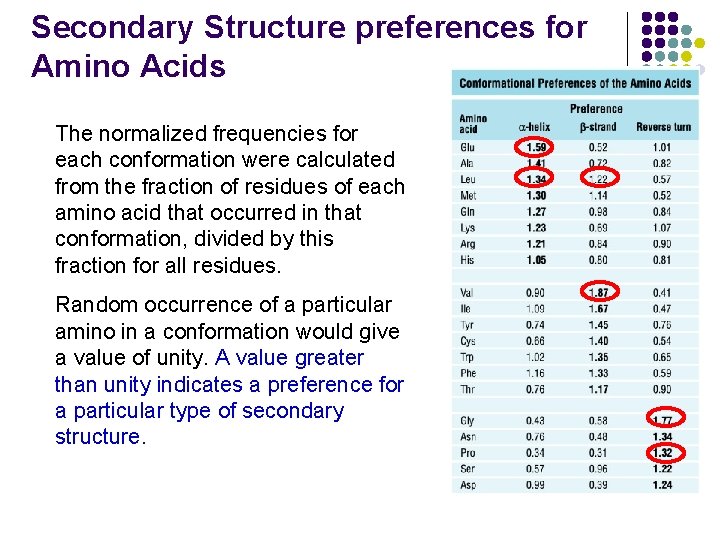
Secondary Structure preferences for Amino Acids The normalized frequencies for each conformation were calculated from the fraction of residues of each amino acid that occurred in that conformation, divided by this fraction for all residues. Random occurrence of a particular amino in a conformation would give a value of unity. A value greater than unity indicates a preference for a particular type of secondary structure.

Outline l Protein Structure l l l Why structure How to Predict Protein Structure l Experimental methods l Computational methods (predictive methods) Protein Structure Prediction l Secondary structure prediction (2 D) l l Machine learning methods for Protein Secondary Structure Prediction Tertiary structure prediction (3 D) l Ab initio l Homology modeling
Machine learning methods for Protein Secondary Structure Prediction l l l Introduction to classification Generalize protein secondary structure prediction as a machine learning problem Introduction to Neural Network
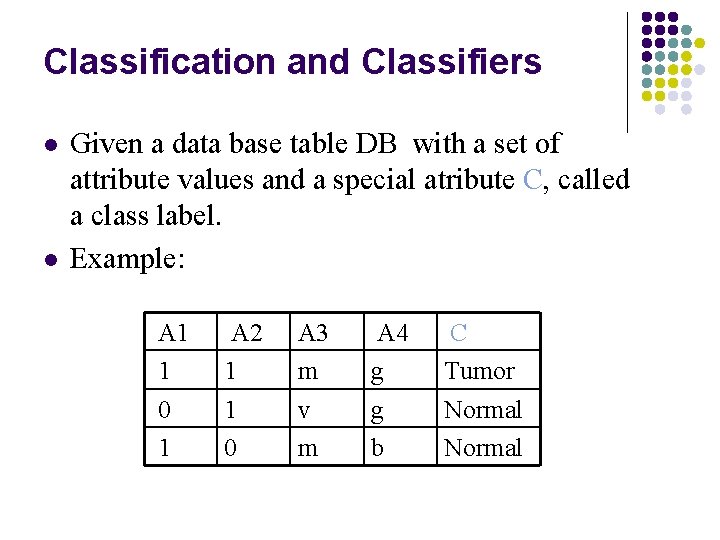
Classification and Classifiers l l Given a data base table DB with a set of attribute values and a special atribute C, called a class label. Example: A 1 1 0 1 A 2 1 1 0 A 3 m v m A 4 g g b C Tumor Normal
Classification and Classifiers l An algorithm is called a classification algorithm if it uses the data to build a set of patterns l l Decision rules or decision trees, etc. Those patters are structured in such a way that we can use them to classify unknown sets of objects- unknown records. For that reason (because of the goal) the classification algorithm is often called shortly a classifier. Classifier Example
Classification and Classifiers l Building a classifier consists of two phases: l l l The training data set to create patterns (rules, trees, or to train a Neural network). l l Training and testing. In both phases we use data (training data set and disjoint test data set) for which the class labels are known for ALL of the records. Evaluate created patterns with the use of of test data, which classification is known. The measure for a trained classifier accuracy is called predictive accuracy.
Predictive Accuracy Evaluation The main methods of predictive accuracy evaluations are: • • Re-substitution (N ; N) Holdout (2 N/3 ; N/3) x-fold cross-validation (N-N/x ; N/x) Leave-one-out (N-1 ; 1), where N is the number of instances in the dataset l The process of building and evaluating a classifier is also called a supervised learning, or lately when dealing with large data bases a classification method in Data Mining
Classification Models: Different Classifiers Typical classification models l Decision Trees (ID 3, C 4. 5) l Nearest Neighbors l Support Vector Machines l Neural Networks l l Most of the best classifiers for PSSP are based on Neural Network model Demonstration
Machine learning methods for Protein Secondary Structure Prediction l l l Introduction to classification Generalize protein secondary structure prediction as a machine learning problem Introduction to Neural Network
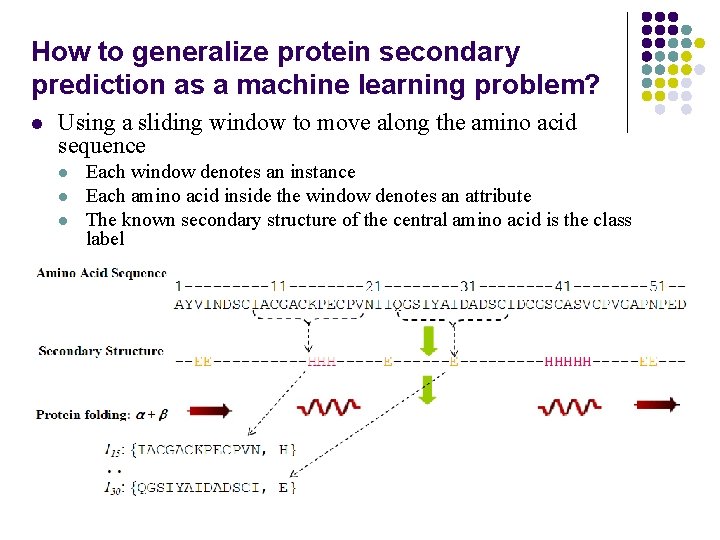
How to generalize protein secondary prediction as a machine learning problem? l Using a sliding window to move along the amino acid sequence l l l Each window denotes an instance Each amino acid inside the window denotes an attribute The known secondary structure of the central amino acid is the class label
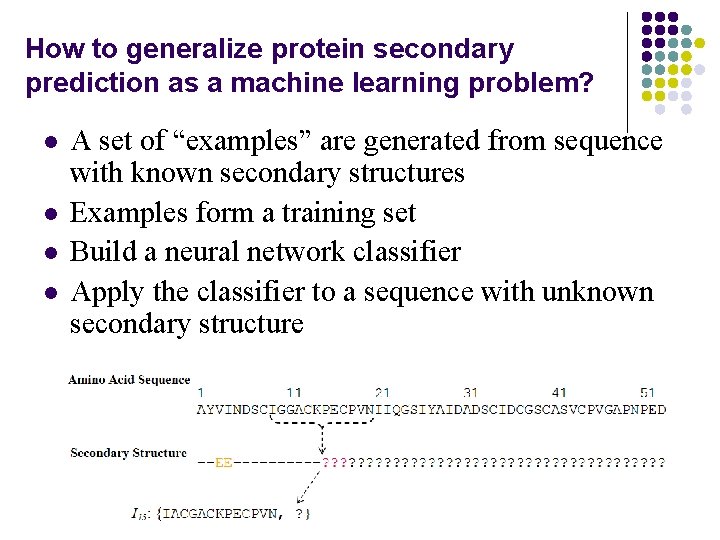
How to generalize protein secondary prediction as a machine learning problem? l l A set of “examples” are generated from sequence with known secondary structures Examples form a training set Build a neural network classifier Apply the classifier to a sequence with unknown secondary structure
Machine learning methods for Protein Secondary Structure Prediction l l l Introduction to classification Generalize protein secondary structure prediction as a machine learning problem Introduction to Neural Network
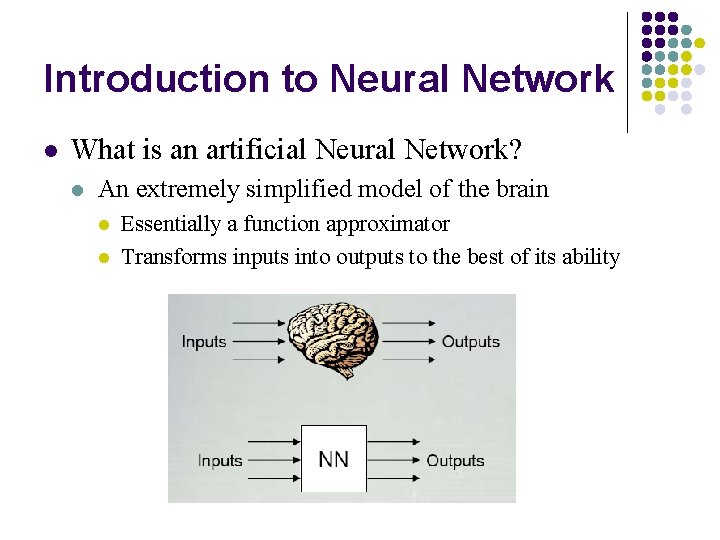
Introduction to Neural Network l What is an artificial Neural Network? l An extremely simplified model of the brain l l Essentially a function approximator Transforms inputs into outputs to the best of its ability
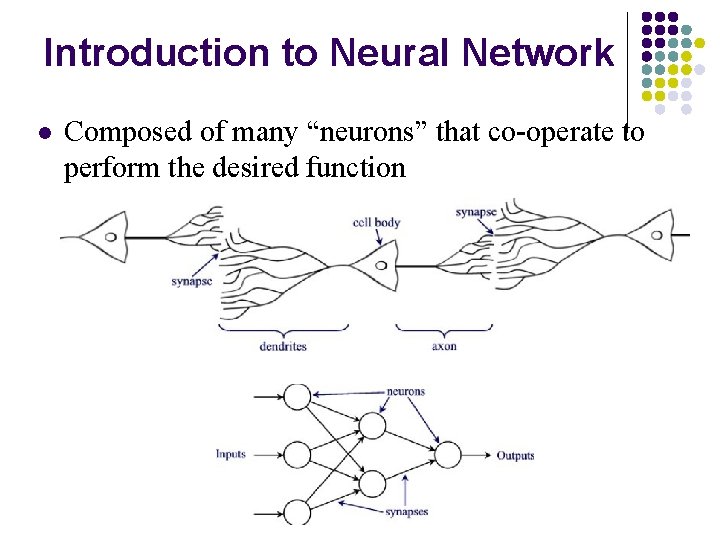
Introduction to Neural Network l Composed of many “neurons” that co-operate to perform the desired function
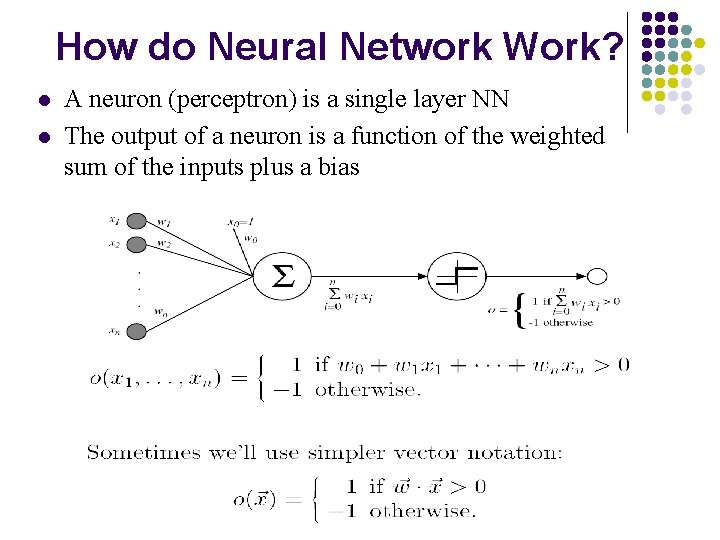
How do Neural Network Work? l l A neuron (perceptron) is a single layer NN The output of a neuron is a function of the weighted sum of the inputs plus a bias

Activation Function l Binary active function l l l f(x)=1 if x>=0 f(x)=0 otherwise The most common sigmoid function used is the logistic function l l f(x) = 1/(1 + e-x) The calculation of derivatives are important for neural networks and the logistic function has a very nice derivative l f’(x) = f(x)(1 - f(x))

Where Do The Weights Come From? l l The weights in a neural network are the most important factor in determining its function Training is the act of presenting the network with some sample data and modifying the weights to better approximate the desired function l Supervised Training l l Supplies the neural network with inputs and the desired outputs Response of the network to the inputs is measured § The weights are modified to reduce the difference between the actual and desired outputs
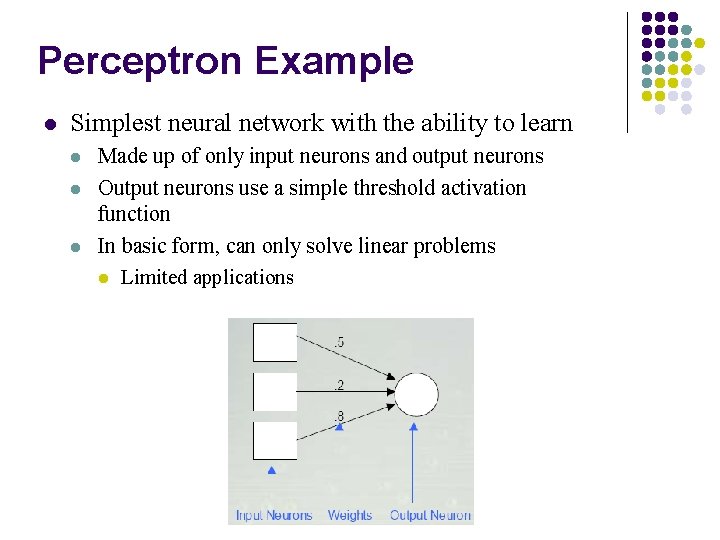
Perceptron Example l Simplest neural network with the ability to learn l l l Made up of only input neurons and output neurons Output neurons use a simple threshold activation function In basic form, can only solve linear problems l Limited applications
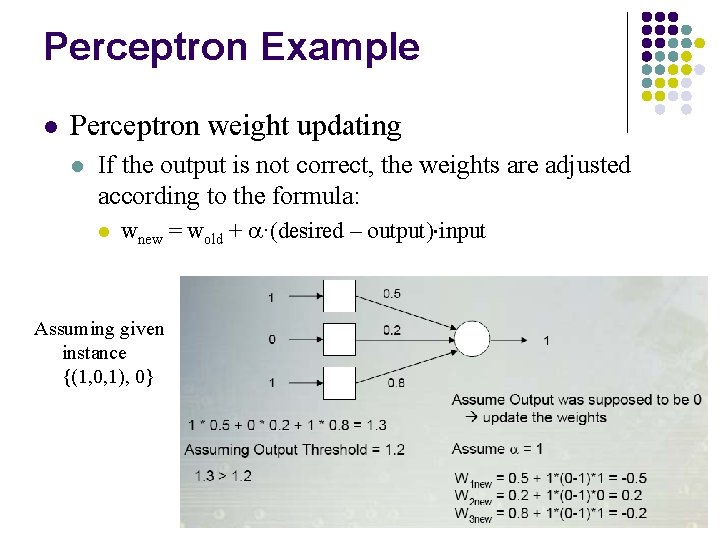
Perceptron Example l Perceptron weight updating l If the output is not correct, the weights are adjusted according to the formula: l wnew = wold + ·(desired – output) input Assuming given instance {(1, 0, 1), 0}
Multi-Layer Feedforward NN l An extension of the perceptron l Multiple layers l l Activation function is not simply a threshold l l Usually a sigmoid function A general function approximator l l The addition of one or more “hidden” layers in between the input and output layers Not limited to linear problems Information flows in one direction l The outputs of one layer act as inputs to the next layer
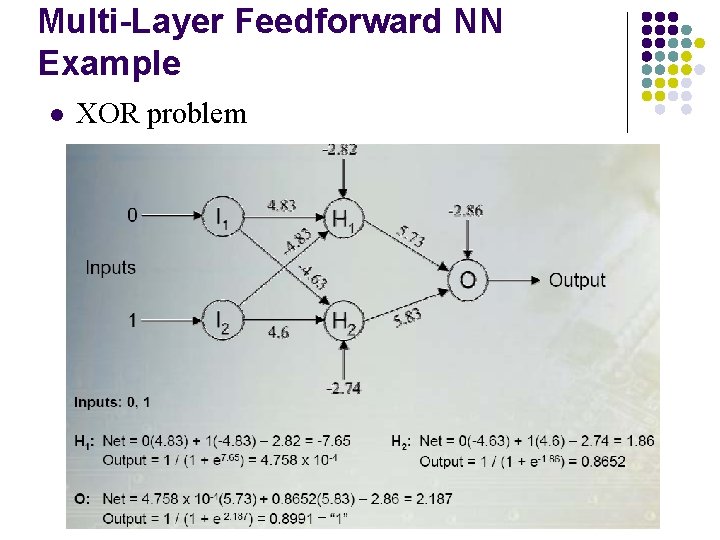
Multi-Layer Feedforward NN Example l XOR problem
Back-propagation l Searches for weight values that minimize the total error of the network over the set of training examples l l Forward pass: Compute the outputs of all units in the network, and the error of the output layers. Backward pass: The network error is used for updating the weights (credit assignment problem).
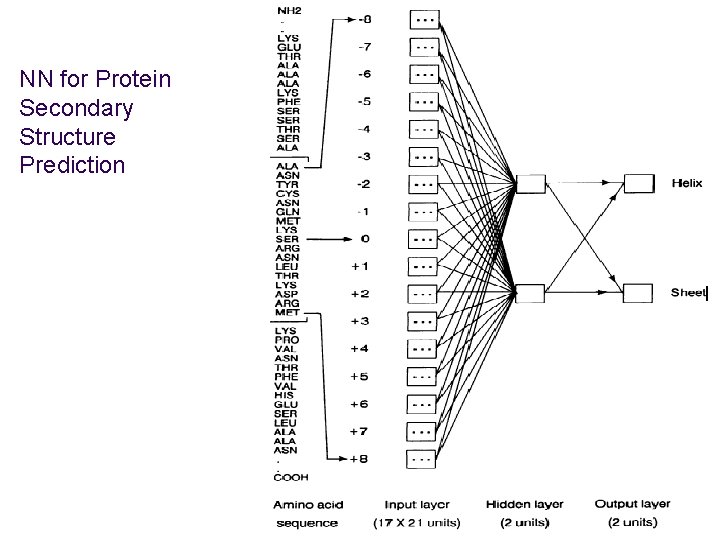
NN for Protein Secondary Structure Prediction

Outline l Protein Structure l l l Why structure How to Predict Protein Structure l Experimental methods l Computational methods (predictive methods) Protein Structure Prediction l Secondary structure prediction (2 D) l l Machine learning methods for Protein Secondary Structure Prediction Tertiary structure prediction (3 D) l Ab initio l Homology modeling
Ab initio Prediction l Sampling the global conformation space l Lattice models / Discrete-state models l Molecular Dynamics l Picking native conformations with an energy function l Solvation model: how protein interacts with water l Pair interactions between amino acids
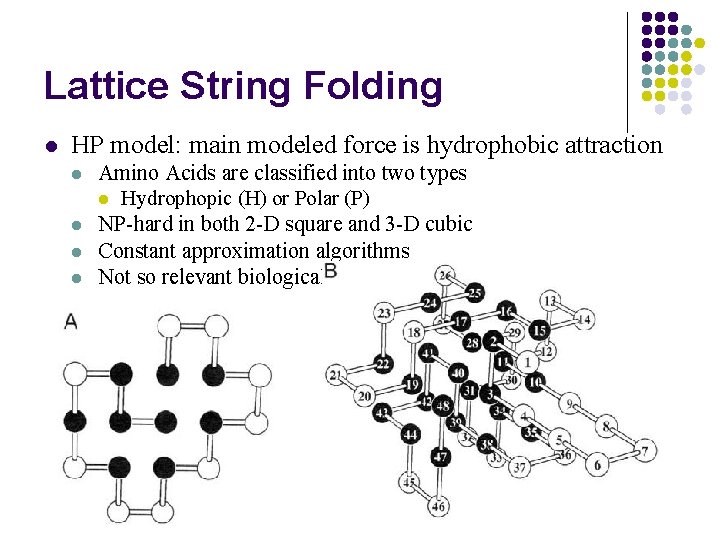
Lattice String Folding l HP model: main modeled force is hydrophobic attraction l l Amino Acids are classified into two types l Hydrophopic (H) or Polar (P) NP-hard in both 2 -D square and 3 -D cubic Constant approximation algorithms Not so relevant biologically
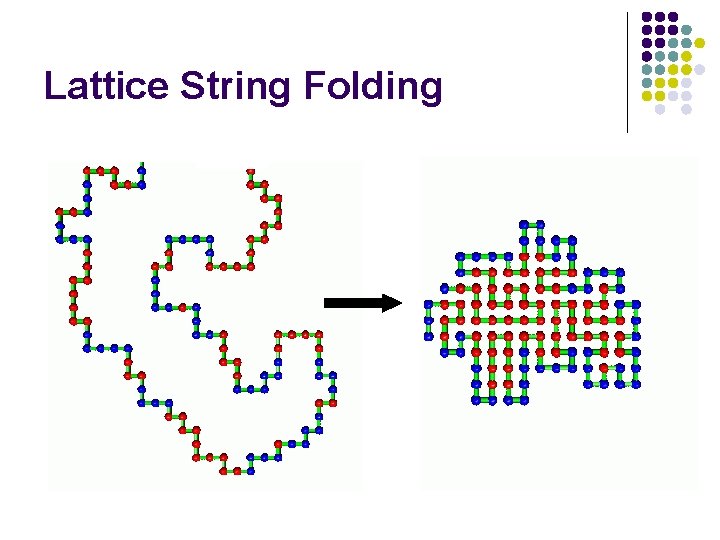
Lattice String Folding

Energy Minimization l Many forces act on a protein l Hydrophobic: inside of protein wants to avoid water l l l Packing: atoms can't be too close, nor too far away van der Waals interactions Bond angle/length constraints Long distance, e. g. l l l Hydrophobic molecules associate with each other in water solvent as if water molecules is the repellent to them. It is like oil/water separation. Electrostatics & Hydrogen bonds Disulphide bonds Salt bridges Can calculate all of these forces, and minimize Intractable in general case, but can be useful
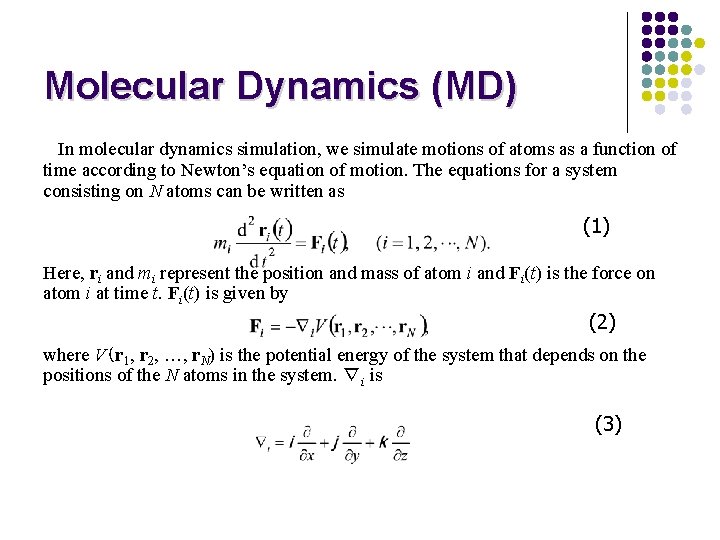
Molecular Dynamics (MD) In molecular dynamics simulation, we simulate motions of atoms as a function of time according to Newton’s equation of motion. The equations for a system consisting on N atoms can be written as (1) Here, ri and mi represent the position and mass of atom i and Fi(t) is the force on atom i at time t. Fi(t) is given by (2) where V(r 1, r 2, …, r. N) is the potential energy of the system that depends on the positions of the N atoms in the system. ∇i is (3)
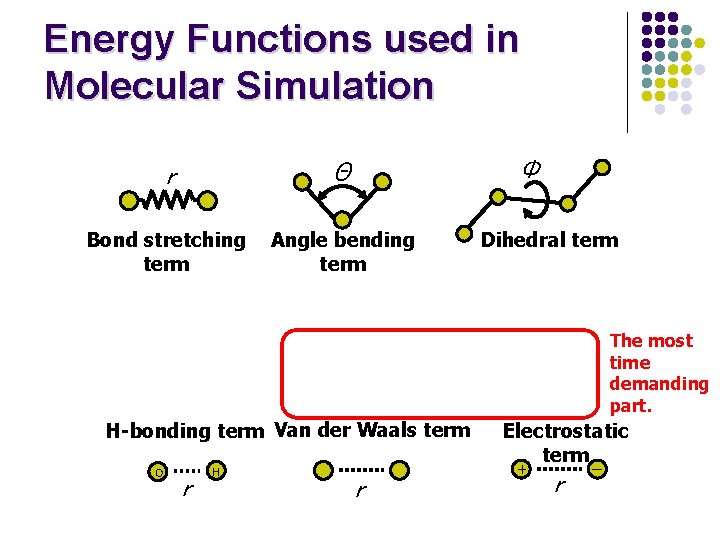
Energy Functions used in Molecular Simulation Φ r Θ Bond stretching term Angle bending term Dihedral term The most time demanding part. H-bonding term Van der Waals term O r H r Electrostatic term + r ー

Outline l Protein Structure l l l Why structure How to Predict Protein Structure l Experimental methods l Computational methods (predictive methods) Protein Structure Prediction l Secondary structure prediction (2 D) l l Machine learning methods for Protein Secondary Structure Prediction Tertiary structure prediction (3 D) l Ab initio l Homology modeling
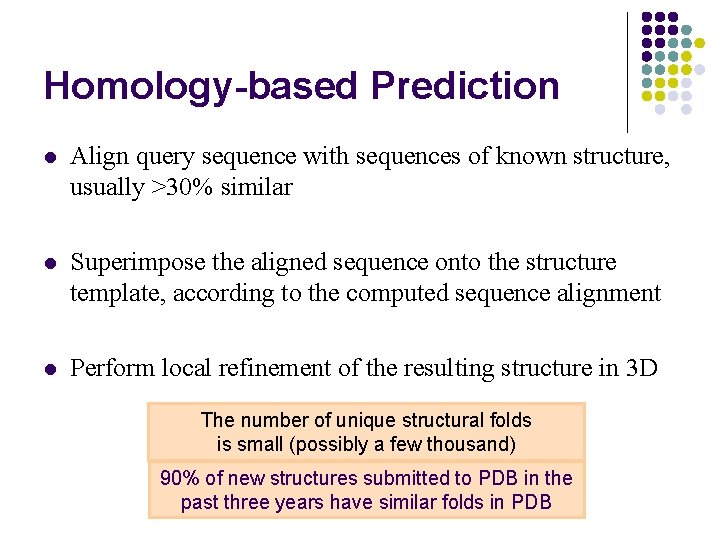
Homology-based Prediction l Align query sequence with sequences of known structure, usually >30% similar l Superimpose the aligned sequence onto the structure template, according to the computed sequence alignment l Perform local refinement of the resulting structure in 3 D The number of unique structural folds is small (possibly a few thousand) 90% of new structures submitted to PDB in the past three years have similar folds in PDB
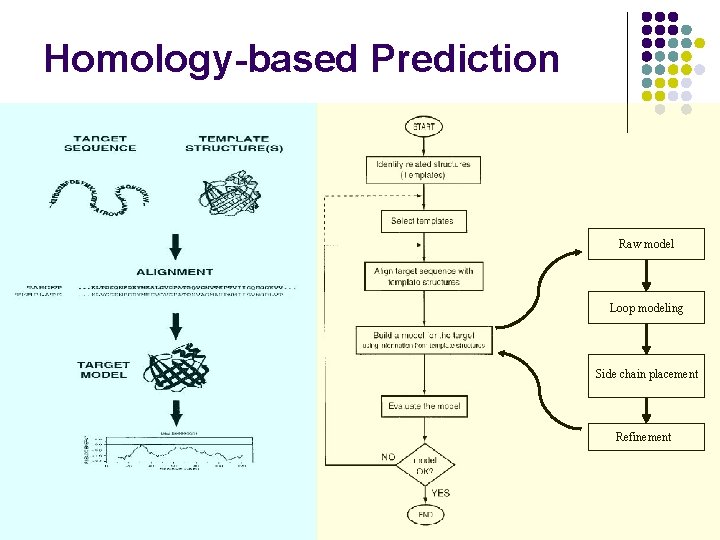
Homology-based Prediction Raw model Loop modeling Side chain placement Refinement
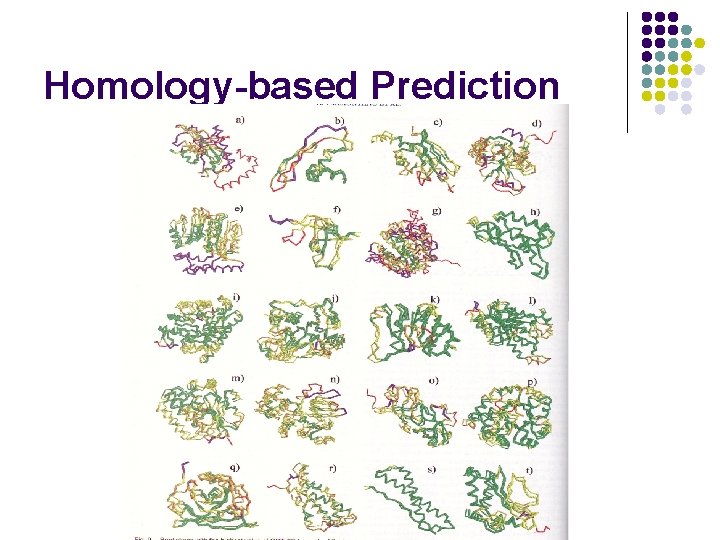
Homology-based Prediction
Outline l Protein Structure l l l Why structure How to predict protein structure l Experimental methods l Computational methods (predictive methods) Protein Structure Prediction l Secondary structure prediction (2 D) l l Machine learning methods for protein secondary structure prediction Tertiary structure prediction (3 D) l Ab initio l Homology modeling
- Slides: 58