Correlating traits with phylogenies Using Ba TS Phylogeny
Correlating traits with phylogenies Using Ba. TS
Phylogeny and trait values l l l A phylogeny describes a hypothesis about the evolutionary relationship between individuals sampled from a population Discrete character traits of interest can be mapped onto the phylogeny A significant association between a particular trait value and its distribution on a phylogeny indicates a potential causative relationship
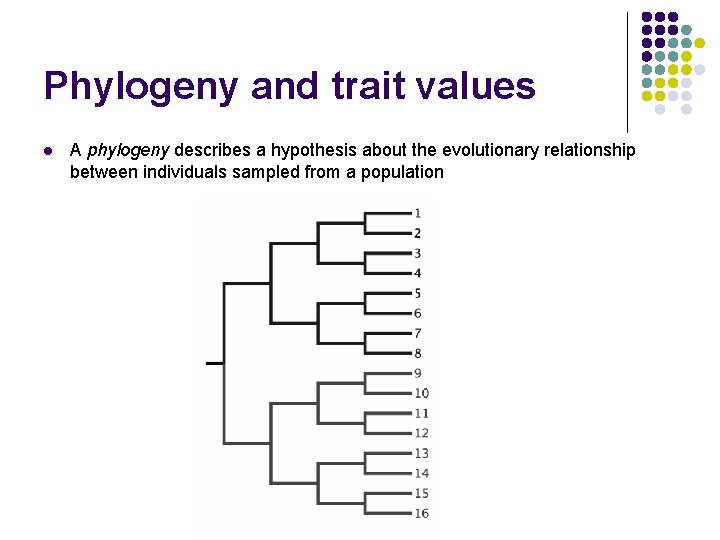
Phylogeny and trait values l A phylogeny describes a hypothesis about the evolutionary relationship between individuals sampled from a population
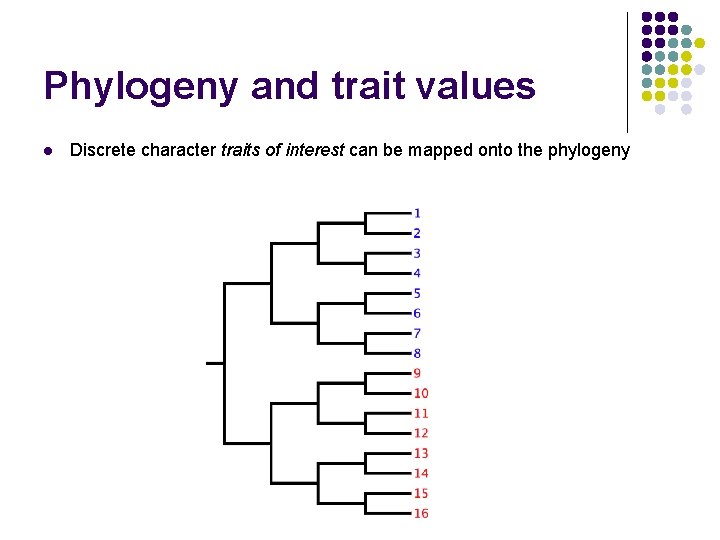
Phylogeny and trait values l Discrete character traits of interest can be mapped onto the phylogeny
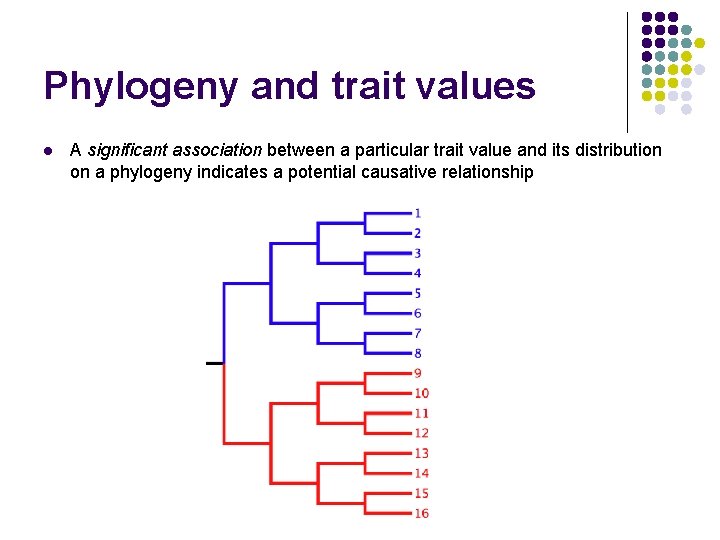
Phylogeny and trait values l A significant association between a particular trait value and its distribution on a phylogeny indicates a potential causative relationship
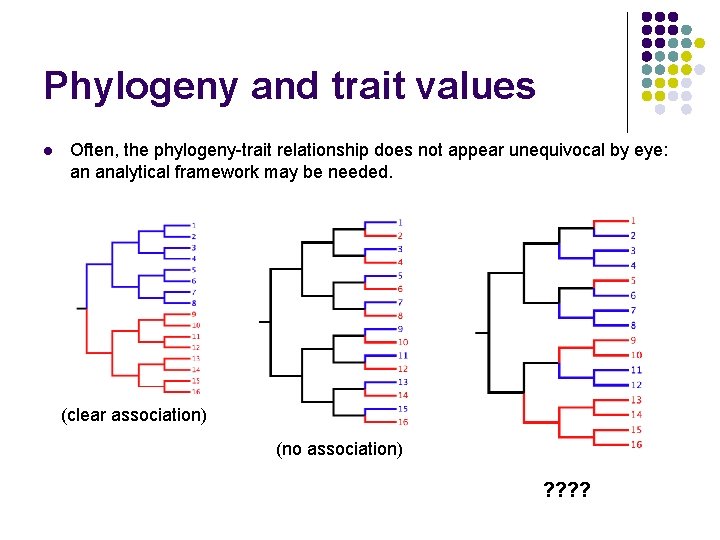
Phylogeny and trait values l Often, the phylogeny-trait relationship does not appear unequivocal by eye: an analytical framework may be needed. (clear association) (no association) ? ?

Phylogeny and trait values The null hypothesis under test is one of random phylogeny-trait association; that is, that “No single tip bearing a given character trait is any more likely to share that trait with adjoining taxa than we would expect due to chance”
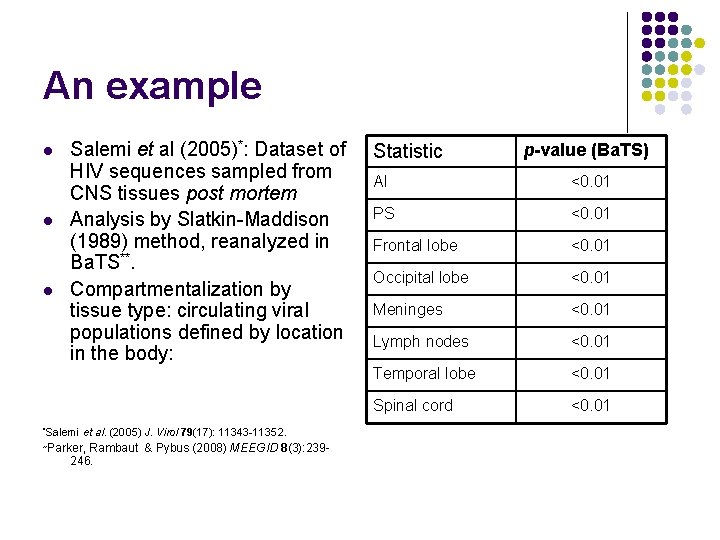
An example l l l Salemi et al (2005)*: Dataset of HIV sequences sampled from CNS tissues post mortem Analysis by Slatkin-Maddison (1989) method, reanalyzed in Ba. TS**. Compartmentalization by tissue type: circulating viral populations defined by location in the body: *Salemi et al. (2005) J. Virol 79(17): 11343 -11352. Parker, Rambaut & Pybus (2008) MEEGID 8(3): 239246. ** Statistic p-value (Ba. TS) AI <0. 01 PS <0. 01 Frontal lobe <0. 01 Occipital lobe <0. 01 Meninges <0. 01 Lymph nodes <0. 01 Temporal lobe <0. 01 Spinal cord <0. 01

Available methods l Non-phylogenetic: ANOVA l l Ignores shared ancestry Phylogenetic: l l l Single tree mapping Slatkin-Maddison & AI Ba. TS

Methods: Single-tree mapping l Method: l l l Pros: l l l Map traits onto a tree Look for correlation Fast Simple Cons: l l l No indication of significance Statistically weak (high Type II error) Conditional on a single topology

Methods: Slatkin-Maddison & AI l Method: l l l Pros: l l l Map traits onto a tree by parsimony & count migration events (Slatkin-Maddison) or measure ‘association index’ within clades recursively (AI) Compare observed value with a null (expected) value obtained by bootstrapping Still reasonably fast Indication of significance Cons: l Still conditional on a single topology
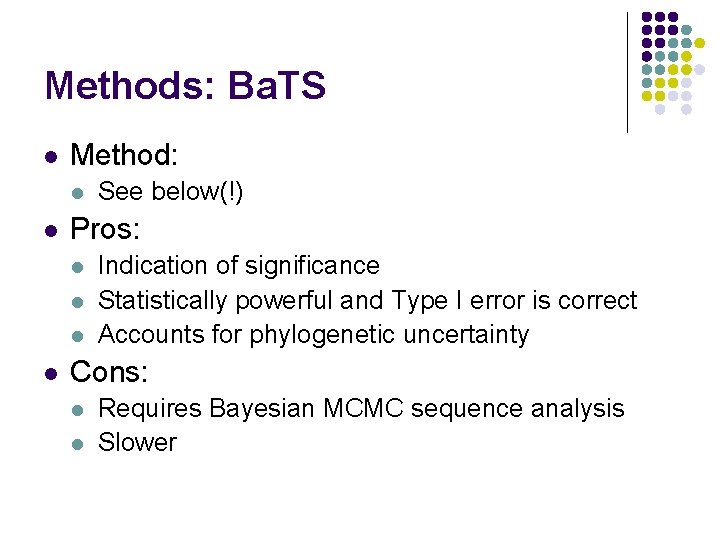
Methods: Ba. TS l Method: l l Pros: l l See below(!) Indication of significance Statistically powerful and Type I error is correct Accounts for phylogenetic uncertainty Cons: l l Requires Bayesian MCMC sequence analysis Slower
Ba. TS: under the bonnet l l Use a posterior distribution of phylogenies from Bayesian MCMC analysis Calculates migrations, AI and a variety of other measures of association Both observed and expected (null) values’ posterior distributions sampled Significance obtained by comparing observed vs. expected
Ba. TS: analysis workflow l Preparation: l l l Sequence alignment Bayesian MCMC phylogeny reconstruction (BEAST, Mr. BAYES) to obtain posterior distribution of trees (PST) Taxa in PST marked up with discrete traits Ba. TS analysis Interpretation

Workflow: Preparation (i) l Sequence alignment: l l Bayesian MCMC analysis: l l CLUSTAL, Bio. Edit, SE-Al MRBAYES, BEAST Taxa marked-up with traits

Workflow: Preparation (ii) l Taxa marked-up with traits: Typical NEXUS format:
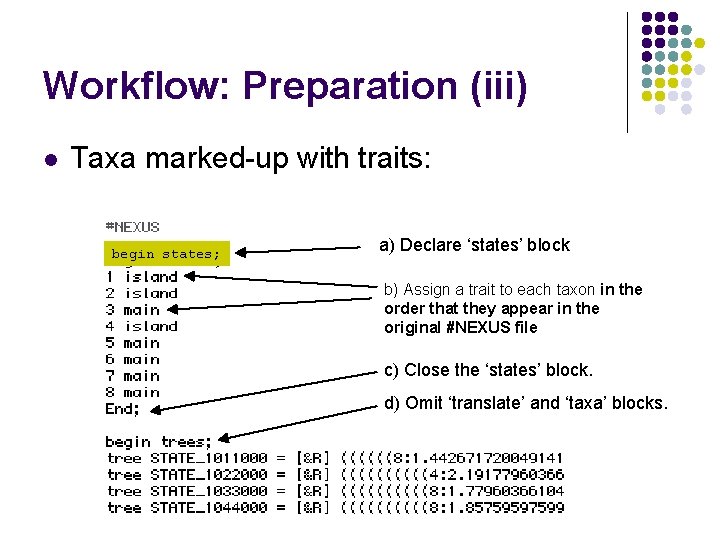
Workflow: Preparation (iii) l Taxa marked-up with traits: begin states; a) Declare ‘states’ block b) Assign a trait to each taxon in the order that they appear in the original #NEXUS file c) Close the ‘states’ block. d) Omit ‘translate’ and ‘taxa’ blocks.
Workflow: Ba. TS analysis To use Ba. TS from the command-line, type: java –jar Ba. TS_beta_build 2. jar [single|batch] <treefile_name> <reps> <states> Where: single or batch asks Ba. TS to analyse either a single input file, or a whole directory (batch analysis) <treefile_name> is the name and full location of the treefile or directory to be analysed, <reps> is the number (an integer > 1, typically 100 at least) of state randomizations to perform to yield a null distribution, and <states> is the number of different states seen.
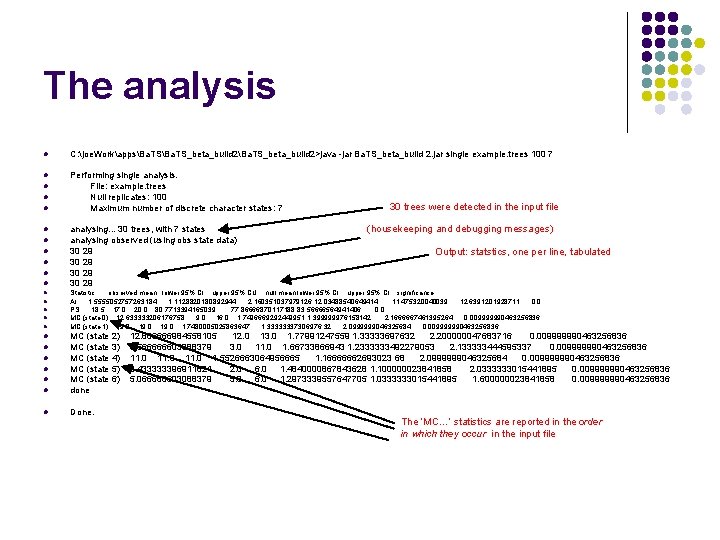
The analysis l C: joe. WorkappsBa. TS_beta_build 2Ba. TS_beta_build 2>java -jar Ba. TS_beta_build 2. jar single example. trees 100 7 l Performing single analysis. File: example. trees Null replicates: 100 Maximum number of discrete character states: 7 l l l l analysing. . . 30 trees, with 7 states analysing observed (using obs state data) 30 29 l l Done. l l (housekeeping and debugging messages) Output: statstics, one per line, tabulated Statistic observed mean lower 95% CI upper 95% CU null mean lower 95% CI upper 95% CI significance AI 1. 5555052757263184 1. 1128820180892944 2. 160351037979126 12. 03488540649414 11. 475320040039 12. 6391201928711 0. 0 PS 18. 5 17. 0 20. 0 80. 7713394165039 77. 86666870117188 83. 56666564941406 0. 0 MC (state 0) 12. 633333206176758 9. 0 16. 0 1. 7496669292449951 1. 399999976158142 2. 1666667461395264 0. 009999990463256836 MC (state 1) 19. 0 1. 7480005025863647 1. 33333337306976 32 2. 0999999046325684 0. 009999990463256836 MC (state 2) MC (state 3) MC (state 4) MC (state 5) MC (state 6) done l 30 trees were detected in the input file 12. 666666984558105 12. 0 13. 0 1. 77991247559 1. 33333697632 2. 200000047683716 0. 009999990463256836 8. 566666603088379 3. 0 11. 0 1. 66733866943 1. 2333333492279053 2. 133333444595337 0. 009999990463256836 11. 0 1. 5526663064956665 1. 16666662693023 68 2. 0999999046325684 0. 009999990463256836 3. 433333396911621 2. 0 6. 0 1. 4840000867843628 1. 100000023841858 2. 0333333015441895 0. 009999990463256836 5. 066666603088379 5. 0 6. 0 1. 2973339557647705 1. 0333333015441895 1. 600000023841858 0. 009999990463256836 The ‘MC…’ statistics are reported in the order in which they occur in the input file

Workflow: Interpretation The null hypothesis under test is one of random phylogeny-trait association; that is, that “No single tip bearing a given character trait is any more likely to share that trait with adjoining taxa than we would expect due to chance”
Workflow: Interpretation The statistics: l Larger values increased phylogeny-trait association l Significance indicated by p-value l In addition, observed posterior values are informative for some statistics: l l PS: indicates migration events between trait values MC(trait value): indicates number of taxon in largest clade monophyletic for that trait value

FAQs / common pitfalls l l Java 1. 5 or higher is required. See java. sun. com for more. Large datasets can be slow, so down-sample input tree files (uniformly, not randomly) where necessary, or to check Ba. TS input files are marked-up correctly. A RAM (memory) shortage can slow the analysis, use –Xmx switch to allocate virtual RAM* Check input file mark-up carefully if in doubt. *See more: http: //edocs. bea. com/wls/docs 70/perform/JVMTuning. html

Author contact: Joe Parker Department of Zoology Oxford University, UK OX 1 3 PS joe@kitserve. org. uk http: //evolve. zoo. ox. ac. uk
- Slides: 23