Correlating PPI node degree with SNP counts Michael














- Slides: 30

Correlating PPI node degree with SNP counts Michael Grobe (This work supported in part by: Research Technologies Indiana University) 1

Do PPI nodes of high degree “have” more or fewer SNPs? Are hubs more or less susceptible to SNPs over evolutionary time? If so, why? If not, why? 2

Hypothesis: The degrees of genes in a PPI network will correlate inversely with their SNP count. This hypothesis will be tested using (parts of) the following data resources: - db. SNP from NCBI, - the Disease Gene Network data collected by Rual, et al. , Stetzl, et al. , and Goh, et al. - several other NCBI resources 3

db. SNP is a large relational database maintained by the National Center for Biotechnology Information (NCBI) on a Microsoft SQLServer. (db. SNP seems to be misnamed. ) NCBI provides several public interfaces to db. SNP: - a web-based interface for public use http: //www. ncbi. nlm. nih. gov/SNP/ - a set of web-accessible scripts CGI scripts and (SOAP-based) Web Services, known as the Entrez e. Utils, and, - an FTP repository of the data exported from the MS SQLServer. NCBI does NOT provide an interface for submitting SQL commands directly to the SQLServer. However, IUSM downloads the db. SNP data from the NCBI FTP repository, loads it into a local MS SQLServer, where it is available for use via JDBC, and UITS makes it available via Web pages and (SOAP-based) Web Services. 4

UITS maintains (on a DB 2 datbase management system) a collection of data resources called the Centralized Life Sciences Data (CLSD) service that incorporates db. SNP via “data federation”. db. SNP can be access via CLSD at http: //discern. uits. iu. edu: 8421/access/index. html and also via a SOAP-based interface to CLSD at http: //discern. uits. iu. edu: 8421/axis/CLSDservice. jws? wsdl db. SNP can also be accessed via JDBC, or through a direct JAX-RPC interface, if necessary. CLSD is described in detail at http: //rac. uits. iu. edu/clsd/ 5

List of CLSD data resources BIND -- Pathways, Gene interactions ENZYME -- Enzyme nomenclature e. PCR -- e. PCR results of Uni. STS vs Homo sapiens SGD -- Saccharomyces Genome Database DGN – The Disease Gene Network data from Goh, et al. (Provisional) KEGG data sources: + LIGAND -- Pathways, Reactions, & Compounds + PATHWAY -- Pathway map coordinates NCBI data sources: + Locus. Link -- Genetic Loci. (retained for archival use. ) + Uni. Gene -- Gene clusters Federated data sources, where the data is stored: * at the originating site: + NCBI Nucleotide -- Nucleotide sequences + NCBI Pub. Med -- Journal abstracts * on local (mirror) servers external to CLSD but housed at IU * BLAST -- Basic Local Alignment Search Tool (mirrored at IU by UITS) * Nucleotide data: NT * Protein data: NR and Swiss-Prot * db. SNP -- Single Nucleotide Polymorphisms (mirrored at IU by IUSM) 6

db. SNP is a relatively complex database. It includes about 300 tables for each species, and the separate species tables share about 80 additional tables. db. SNP is also rather large: db. SNP catalogs Shared, Human, and Mouse (circa early 2008) fill around 150 GB and 3 billion rows (of which about 2. 8 billion are in db. SNP 128_human). New versions come out every 6 months or so. This study uses Build 128, although Build 129 has been quite recently announced. The tutorial “Using db. SNP via SQL queries” describes the structure and use of db. SNP via SQL. 7

The DGN PPI network The Disease Gene Network data within CLSD includes 3 networks: - a network of diseases that are “connected” when they involve the same gene, - a network of 1777 genes that are “connected” when they are implicated in the same disease, and - a Protein Interaction (PPI) network built from networks defined by two different groups Rual, et al. and Stelzl, et al. The PPI is defined in the table called PPI_RUAL_STELZL; it has 7533 unique genes and 22, 052 edges (in a half-matrix form). A companion table PPI_GENES lists every gene in the PPI network The PPI network was traversed to construct a list of shortest paths from each node to each other node: PPI_SHORTEST_PATH_LENGTHS. This is a kind of transitive closure and contains about 53 M records. 8

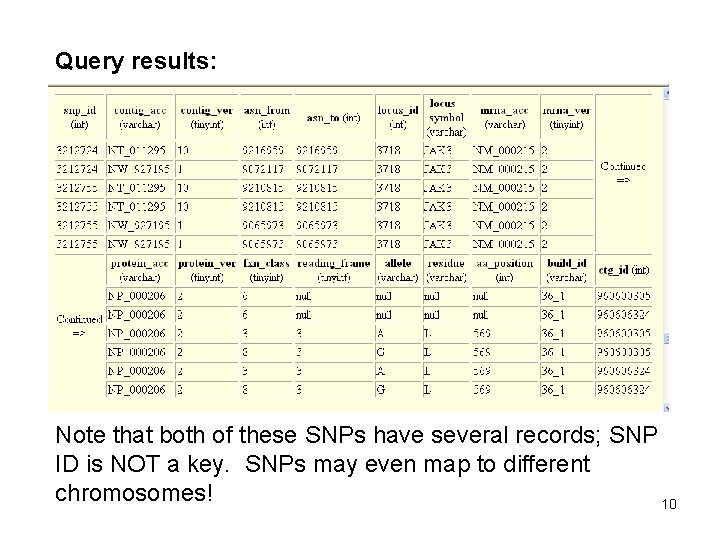
SNPContig. Locus. ID The main db. SNP table used in this project is SNPContig. Locus. ID which contains information about the genes associated with each SNP. The Build 128 version of SNPContig. Locus. ID contains about 13, 129, 868 rows (though about half of them specify “NW_” m. RNA segments and were ignored). Here is a query that retrieves the records for 2 SNPs (among many others) that appear within, or close to, the coding region for JAK 3. select * from b 126_SNPContig. Locus. Id_36_1 where snp_id in ( 3212724, 3212755 ) 9

Query results: Note that both of these SNPs have several records; SNP ID is NOT a key. SNPs may even map to different chromosomes! 10

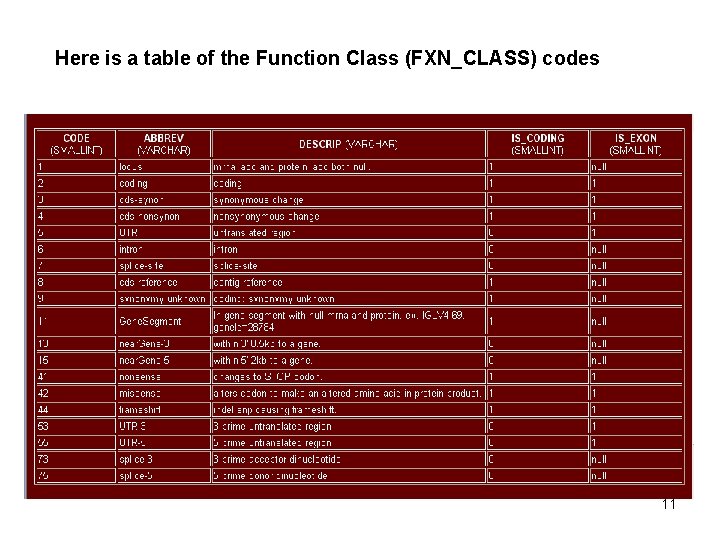
Here is a table of the Function Class (FXN_CLASS) codes . 11

Number of (NT_) SNPs in each SNP function class select fxn_class, count(*) from db. SNP 128_human. b 128_SNPContig. Locus. Id_36_2 where contig_acc like 'NT_%‘ [so not all 13 Mrows will appear] GROUP BY fxn_class ORDER BY fxn_class FXN_CLASS Count 3 78797 6 6008473 8 192868 13 168608 15 166205 41 2753 FXN_CLASS Count 42 98053 44 15848 53 144123 55 27990 73 645 75 483 12

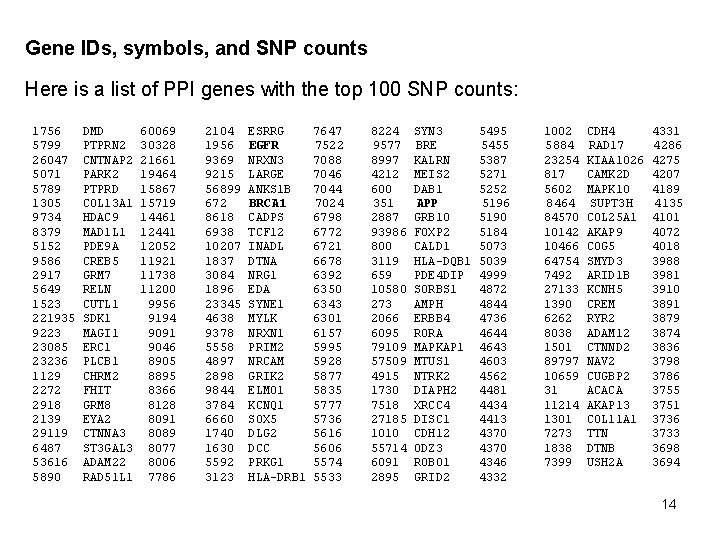
Get gene IDs, symbols, and SNP counts The following query uses both DGN and db. SNP data to get a list of gene IDs, their symbols, and the number SNPs associated with each gene: select a. locus_id, b. locus_symbol, snp_counter from (select locus_id, count(*) as snp_counter from dbsnp 128_human. b 128_SNPContig. Locus. Id_36_2 where contig_acc like 'NT_%' and locus_id in (select gene_id from disease_gene_net. ppi_genes ) group by locus_id) as a join (select distinct locus_id, locus_symbol from dbsnp 128_human. b 128_SNPContig. Locus. Id_36_2) as b on b. locus_id = a. locus_id order by snp_counter desc 13

Gene IDs, symbols, and SNP counts Here is a list of PPI genes with the top 100 SNP counts: 1756 5799 26047 5071 5789 1305 9734 8379 5152 9586 2917 5649 1523 221935 9223 23085 23236 1129 2272 2918 2139 29119 6487 53616 5890 DMD PTPRN 2 CNTNAP 2 PARK 2 PTPRD COL 13 A 1 HDAC 9 MAD 1 L 1 PDE 9 A CREB 5 GRM 7 RELN CUTL 1 SDK 1 MAGI 1 ERC 1 PLCB 1 CHRM 2 FHIT GRM 8 EYA 2 CTNNA 3 ST 3 GAL 3 ADAM 22 RAD 51 L 1 60069 30328 21661 19464 15867 15719 14461 12441 12052 11921 11738 11200 9956 9194 9091 9046 8905 8895 8366 8128 8091 8089 8077 8006 7786 2104 1956 9369 9215 56899 672 8618 6938 10207 1837 3084 1896 23345 4638 9378 5558 4897 2898 9844 3784 6660 1740 1630 5592 3123 ESRRG EGFR NRXN 3 LARGE ANKS 1 B BRCA 1 CADPS TCF 12 INADL DTNA NRG 1 EDA SYNE 1 MYLK NRXN 1 PRIM 2 NRCAM GRIK 2 ELMO 1 KCNQ 1 SOX 5 DLG 2 DCC PRKG 1 HLA-DRB 1 7647 7522 7088 7046 7044 7024 6798 6772 6721 6678 6392 6350 6343 6301 6157 5995 5928 5877 5835 5777 5736 5616 5606 5574 5533 8224 9577 8997 4212 600 351 2887 93986 800 3119 659 10580 273 2066 6095 79109 57509 4915 1730 7518 27185 1010 55714 6091 2895 SYN 3 BRE KALRN MEIS 2 DAB 1 APP GRB 10 FOXP 2 CALD 1 HLA-DQB 1 PDE 4 DIP SORBS 1 AMPH ERBB 4 RORA MAPKAP 1 MTUS 1 NTRK 2 DIAPH 2 XRCC 4 DISC 1 CDH 12 ODZ 3 ROBO 1 GRID 2 5495 5455 5387 5271 5252 5196 5190 5184 5073 5039 4999 4872 4844 4736 4644 4643 4603 4562 4481 4434 4413 4370 4346 4332 1002 5884 23254 817 5602 8464 84570 10142 10466 64754 7492 27133 1390 6262 8038 1501 89797 10659 31 11214 1301 7273 1838 7399 CDH 4 RAD 17 KIAA 1026 CAMK 2 D MAPK 10 SUPT 3 H COL 25 A 1 AKAP 9 COG 5 SMYD 3 ARID 1 B KCNH 5 CREM RYR 2 ADAM 12 CTNND 2 NAV 2 CUGBP 2 ACACA AKAP 13 COL 11 A 1 TTN DTNB USH 2 A 4331 4286 4275 4207 4189 4135 4101 4072 4018 3981 3910 3891 3879 3874 3836 3798 3786 3755 3751 3736 3733 3698 3694 14

PPI node degree Here is a query that uses PPI_SHORTEST_PATH_LENGTHS to get degree for each node: select source, count(*) as degree from disease_gene_net. PPI_SHORTEST_PATH_LENGTHS where length = 1 and source in ( select gene_id from DISEASE_GENE_NET. PPI_GENES ) group by source order by degree 15

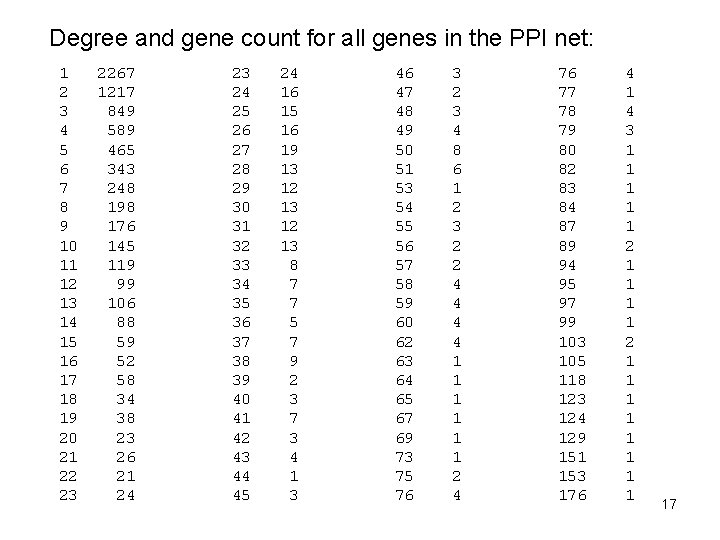
PPI node degree Here is a query using that closure to get gene counts for each degree: select degree, count(*) from (select source, count(*) as degree from disease_gene_net. PPI_SHORTEST_PATH_LENGTHS where length = 1 and source in ( select gene_id from DISEASE_GENE_NET. PPI_GENES ) group by source ) as a group by degree order by degree 16

Degree and gene count for all genes in the PPI net: 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 2267 1217 849 589 465 343 248 198 176 145 119 99 106 88 59 52 58 34 38 23 26 21 24 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 24 16 15 16 19 13 12 13 8 7 7 5 7 9 2 3 7 3 4 1 3 46 47 48 49 50 51 53 54 55 56 57 58 59 60 62 63 64 65 67 69 73 75 76 3 2 3 4 8 6 1 2 3 2 2 4 4 1 1 1 2 4 76 77 78 79 80 82 83 84 87 89 94 95 97 99 103 105 118 123 124 129 151 153 176 4 1 4 3 1 1 1 2 1 1 17

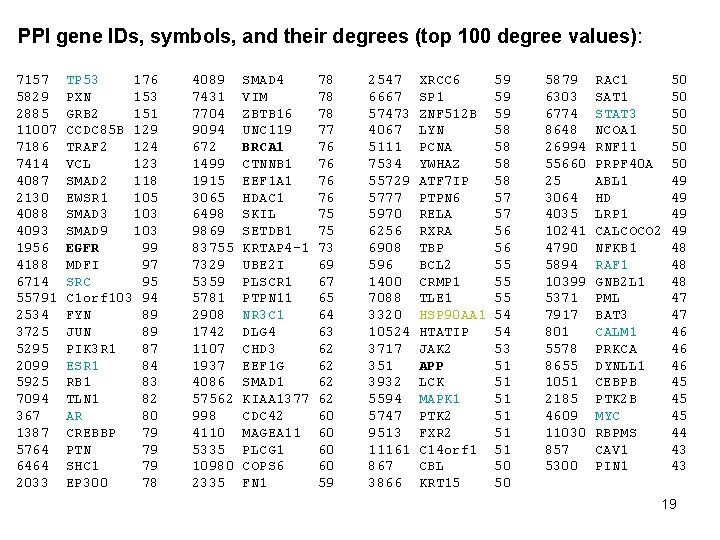
To get PPI gene IDs, symbols, and degrees: select b. locus_id, b. locus_symbol, degree from (select source, count(*) as degree from disease_gene_net. PPI_SHORTEST_PATH_LENGTHS where length = 1 and source in ( select gene_id from DISEASE_GENE_NET. PPI_GENES ) group by source) as a join (select distinct locus_id, locus_symbol from dbsnp 128_human. b 128_SNPContig. Locus. Id_36_2) as b on b. locus_id = a. source 18

PPI gene IDs, symbols, and their degrees (top 100 degree values): 7157 5829 2885 11007 7186 7414 4087 2130 4088 4093 1956 4188 6714 55791 2534 3725 5295 2099 5925 7094 367 1387 5764 6464 2033 TP 53 176 PXN 153 GRB 2 151 CCDC 85 B 129 TRAF 2 124 VCL 123 SMAD 2 118 EWSR 1 105 SMAD 3 103 SMAD 9 103 EGFR 99 MDFI 97 SRC 95 C 1 orf 103 94 FYN 89 JUN 89 PIK 3 R 1 87 ESR 1 84 RB 1 83 TLN 1 82 AR 80 CREBBP 79 PTN 79 SHC 1 79 EP 300 78 4089 7431 7704 9094 672 1499 1915 3065 6498 9869 83755 7329 5359 5781 2908 1742 1107 1937 4086 57562 998 4110 5335 10980 2335 SMAD 4 VIM ZBTB 16 UNC 119 BRCA 1 CTNNB 1 EEF 1 A 1 HDAC 1 SKIL SETDB 1 KRTAP 4 -1 UBE 2 I PLSCR 1 PTPN 11 NR 3 C 1 DLG 4 CHD 3 EEF 1 G SMAD 1 KIAA 1377 CDC 42 MAGEA 11 PLCG 1 COPS 6 FN 1 78 78 78 77 76 76 75 75 73 69 67 65 64 63 62 62 60 60 59 2547 6667 57473 4067 5111 7534 55729 5777 5970 6256 6908 596 1400 7088 3320 10524 3717 351 3932 5594 5747 9513 11161 867 3866 XRCC 6 SP 1 ZNF 512 B LYN PCNA YWHAZ ATF 7 IP PTPN 6 RELA RXRA TBP BCL 2 CRMP 1 TLE 1 HSP 90 AA 1 HTATIP JAK 2 APP LCK MAPK 1 PTK 2 FXR 2 C 14 orf 1 CBL KRT 15 59 59 59 58 58 57 57 56 56 55 55 55 54 54 53 51 51 51 50 50 5879 6303 6774 8648 26994 55660 25 3064 4035 10241 4790 5894 10399 5371 7917 801 5578 8655 1051 2185 4609 11030 857 5300 RAC 1 SAT 1 STAT 3 NCOA 1 RNF 11 PRPF 40 A ABL 1 HD LRP 1 CALCOCO 2 NFKB 1 RAF 1 GNB 2 L 1 PML BAT 3 CALM 1 PRKCA DYNLL 1 CEBPB PTK 2 B MYC RBPMS CAV 1 PIN 1 50 50 50 49 49 48 48 48 47 47 46 46 46 45 45 45 44 43 43 19

Get SNP counts and degree values for each gene in the PPI: select locus_id, degree, snp_counter from (select locus_id, count(*) as snp_counter from dbsnp 128_human. b 128_SNPContig. Locus. Id_36_2 where contig_acc like 'NT_%' and ( fxn_class = 41 or fxn_class = 42 or fxn_class = 44 ) group by locus_id) as a join (select source, count(*) as degree from disease_gene_net. PPI_SHORTEST_PATH_LENGTHS where length = 1 and source in ( select gene_id from DISEASE_GENE_NET. PPI_GENES ) group by source ) as b on source = locus_id order by degree 20

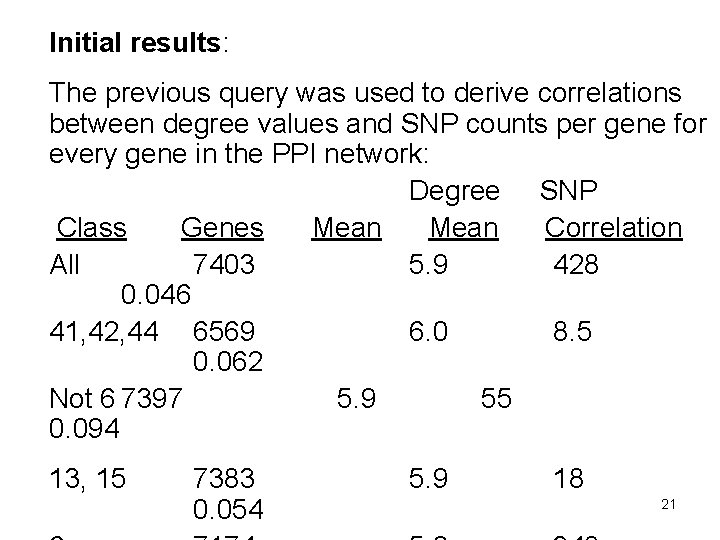
Initial results: The previous query was used to derive correlations between degree values and SNP counts per gene for every gene in the PPI network: Degree SNP Class Genes Mean Correlation All 7403 5. 9 428 0. 046 41, 42, 44 6569 6. 0 8. 5 0. 062 Not 6 7397 5. 9 55 0. 094 13, 15 7383 0. 054 5. 9 18 21

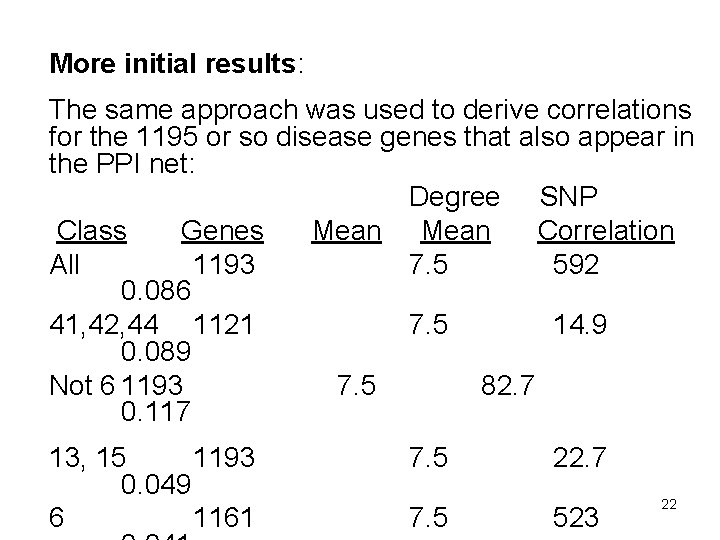
More initial results: The same approach was used to derive correlations for the 1195 or so disease genes that also appear in the PPI net: Degree SNP Class Genes Mean Correlation All 1193 7. 5 592 0. 086 41, 42, 44 1121 7. 5 14. 9 0. 089 Not 6 1193 7. 5 82. 7 0. 117 13, 15 1193 0. 049 6 1161 7. 5 22. 7 523 22

Perhaps a correlation can be found as a function of mer counts? That is, perhaps: “DNA bases in the gene per SNP” or “RNA bases in the gene transcript per SNP” or “amino acids in the protein product per SNP” will correlate with degree, especially for certain SNP classes? Testing these claims requires gene, m. RNA transcript, and/or protein product lengths (and maybe intron lengths). Note that the SNPContig. Locus. Id table includes pointers to m. RNA and protein records, and includes the NCBI UIDs for each record. Scripts (get-m. RNA-lengths. pl and get-protein-lengths. pl) were written to access the m. RNA and protein contig data from NCBI and to count base pairs or amino acids, respectively. 23

Scripts to download mer (base and aa) data The libwww-perl (LWP) module was used to interact with the NCBI e. Utils that were mentioned earlier and are documented in “Using the NCBI e. Utilities via CGI” at http: //mypage. iu. edu/~dgrobe/entrez-dogma. html DNA lengths were obtained using a service at http: //discern. uits. iu. edu: 8421/view-sequences. html called “Get NCBI sequences for genes or specified regions” that will fetch gene FASTA records given gene names and/or NCBI UIDs. NCBI asks users to limit access to one every 3 seconds during off-peak hours and one every 15 seconds otherwise. As a result, these runs took over 24 hours. 24

The resulting “mer file” sizes are like: - DNA length records: - m. RNA length records: - Protein length records: 22259 32400 23803 There are frequently multiple m. RNA and protein records for a gene; mean lengths were computed for each gene by downstream scripts. A script (get-gene-m. RNA-SNPs-mers-per. SNP. pl) was written to compute mean lengths and perform correlations on the mer data. 25

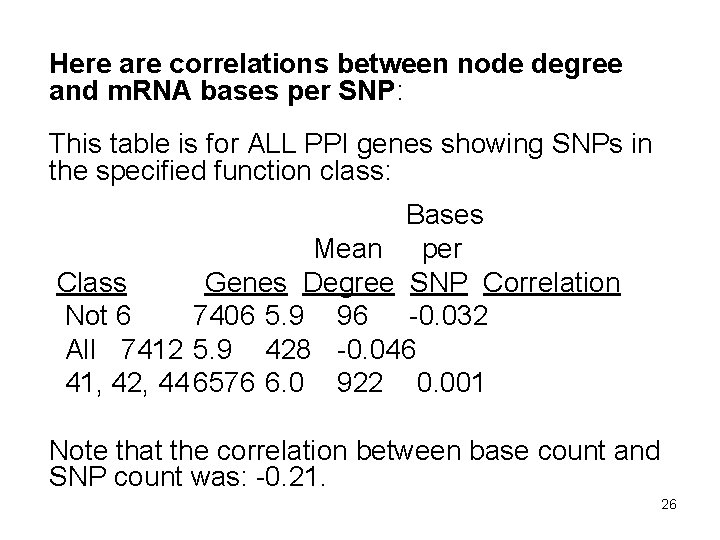
Here are correlations between node degree and m. RNA bases per SNP: This table is for ALL PPI genes showing SNPs in the specified function class: Bases Mean per Class Genes Degree SNP Correlation Not 6 7406 5. 9 96 -0. 032 All 7412 5. 9 428 -0. 046 41, 42, 44 6576 6. 0 922 0. 001 Note that the correlation between base count and SNP count was: -0. 21. 26

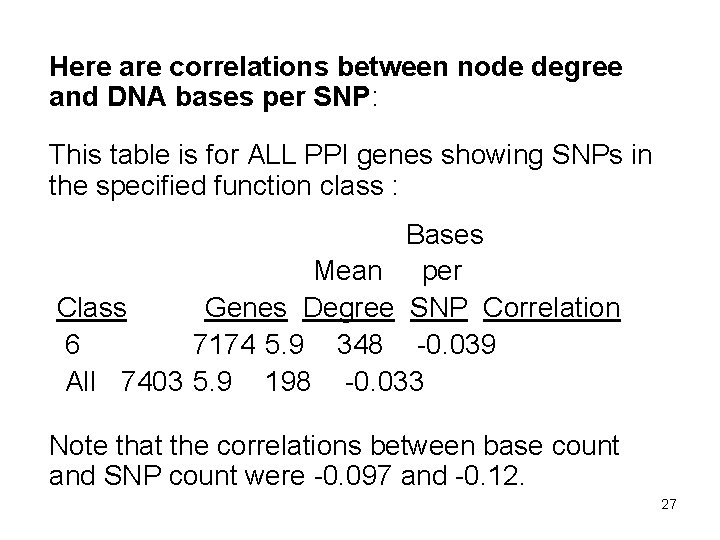
Here are correlations between node degree and DNA bases per SNP: This table is for ALL PPI genes showing SNPs in the specified function class : Bases Mean per Class Genes Degree SNP Correlation 6 7174 5. 9 348 -0. 039 All 7403 5. 9 198 -0. 033 Note that the correlations between base count and SNP count were -0. 097 and -0. 12. 27

Conclusion This study found no relationship between SNP count and PPI node degree, or between measures of mer counts per SNP and node degree. 28

Discussion Are cell networks so robust that variation which “should” normally disrupt functioning gets over-ridden? If so, how? Are there parallel/redundant pathways for important processes? Are non-parallel pathways constructed to minimize the effects of variation? Do chaperone proteins (like HSP 90) help make variant proteins safe for use within the cell (a la’ Whitesell and Lundquist)? (Note: around 20% of HSP-connected genes appear in the list of 100 genes (< 2%) with the highest degree. ) Would hub genes within Reaction networks (as opposed to PPI networks) show SNP counts that correlate with their degree? Would PPIs composed only of co-located proteins display node degree-SNP count correlations? Do lethal genes show fewer SNPs? 29

References The db. SNP Build Process http: //www. ncbi. nlm. nih. gov/books/bookres. fcgi/helpsnpfaq/Build. pdf Using db. SNP via SQL queries http: //mypage. iu. edu/~dgrobe/db. SNP/using-db. SNP-via-SQL. html Using the relational and e. Utils interfaces to db. SNP http: //mypage. iu. edu/~dgrobe/db. SNP/using-db. SNP-at-IU. html Using the NCBI e. Utilities via CGI http: //mypage. iu. edu/~dgrobe/entrez-dogma. html Kwang-Il Goh, Michael E. Cusick David Valle Hum, Barton Childs Hum, Marc Vidal, and Albert-Laszlo Barabasi, The human disease network, PNAS, May 22, 2007, vol. 104, no. 21, 8685. http: //www. pnas. org/content/104/21/8685. abstract Get contents of tables related to the Goh (2007) paper http: //discern. uits. iu. edu: 8421/show-a-DISEASE_GENE_NET-Table. html Whitesell, Luke, and Susan L. Lundquist, HSP and the chaparoning of cancer, Nat Rev Cancer, 2005; 510: 761 -772. 30