corbett LIFE SCIENCE www corbettlifescience com Assay Design

corbett LIFE SCIENCE www. corbettlifescience. com Assay Design for Real-Time PCR 1

corbett LIFE SCIENCE www. corbettlifescience. com Traditional PCR assay design Originated with gel-based detection methods Best guess of primers equalise length and GC content Longer amplicons look better on a gel so typical designs of 300– 500 bp Iterative optimisation of several parameters Mg. Cl 2, annealing temp, extension times, etc Philosophy: fit reaction conditions to the amplicon 2
corbett LIFE SCIENCE www. corbettlifescience. com q. PCR assay design Philosophy: fit amplicon to fixed reaction conditions Standardize reaction chemistry Mg. Cl 2, d. NTPs, salts, enzyme, etc Standardize cycling conditions typically 2 -step; 95°C/60°C Follow design guidelines and use software tools Some chemistries also standardize primer/probe concs. Little or no optimisation needed All assays are handled identically “Plug-and-Play” assay modules 3
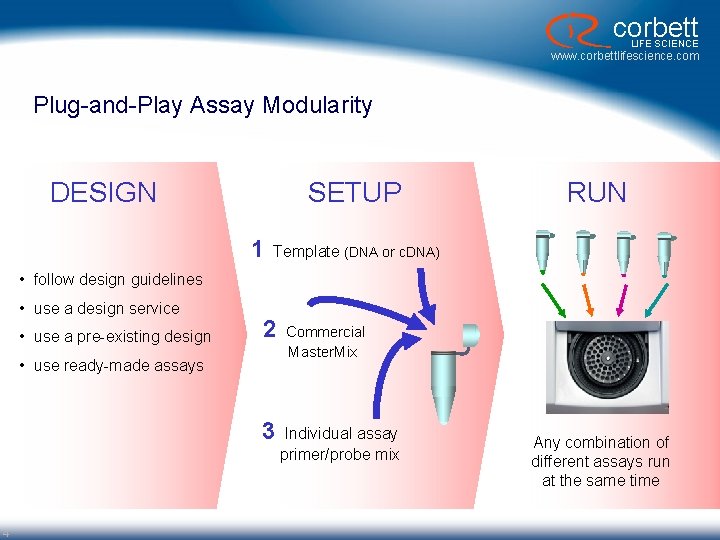
corbett LIFE SCIENCE www. corbettlifescience. com Plug-and-Play Assay Modularity DESIGN SETUP 1 RUN Template (DNA or c. DNA) • follow design guidelines • use a design service • use a pre-existing design 2 • use ready-made assays 3 4 Commercial Master. Mix Individual assay primer/probe mix Any combination of different assays run at the same time
corbett LIFE SCIENCE www. corbettlifescience. com Designing modular assays 1. Define the target sequence 2. Amplicon design guidelines 3. Primer design guidelines 4. Specific chemistry considerations for different probe types etc 5

corbett LIFE SCIENCE www. corbettlifescience. com 1. Define the target sequence understanding the target sequence is critical to success Avoid unusual sequence aim for 40– 60% GC avoid SNPs, pseudogenes & palindromes avoid sequence repeats or repetitive elements avoid discrepancies between data sources Tailor specificity regions of sequence conservation for broad specificity target subspecies by particular base variations 6

corbett LIFE SCIENCE www. corbettlifescience. com http: //restools. sdsc. edu/biotools 16. html#Part 3 7
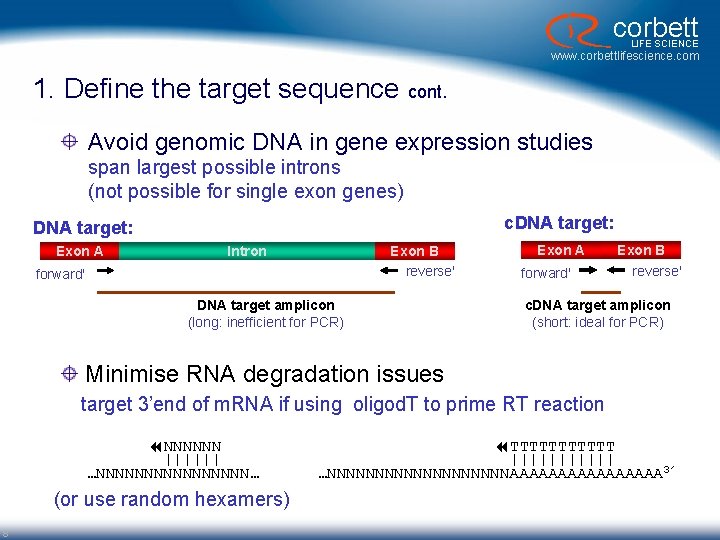
corbett LIFE SCIENCE www. corbettlifescience. com 1. Define the target sequence cont. Avoid genomic DNA in gene expression studies span largest possible introns (not possible for single exon genes) c. DNA target: Exon A Intron Exon B reverse' forward' DNA target amplicon (long: inefficient for PCR) Exon A forward' Exon B reverse' c. DNA target amplicon (short: ideal for PCR) Minimise RNA degradation issues target 3’end of m. RNA if using oligod. T to prime RT reaction NNNNNN |||||| …NNNNNNNN… (or use random hexamers) 8 TTTTTT |||||| …NNNNNNNNNNAAAAAAAA 3’
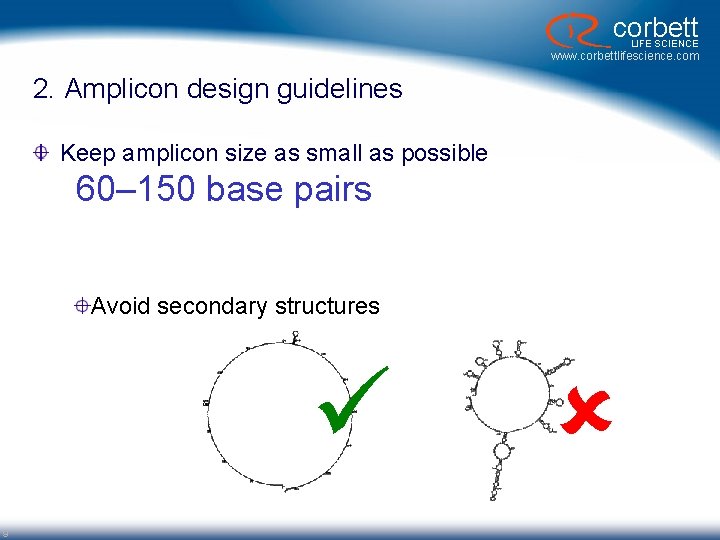
corbett LIFE SCIENCE www. corbettlifescience. com 2. Amplicon design guidelines Keep amplicon size as small as possible 60– 150 base pairs Avoid secondary structures 9
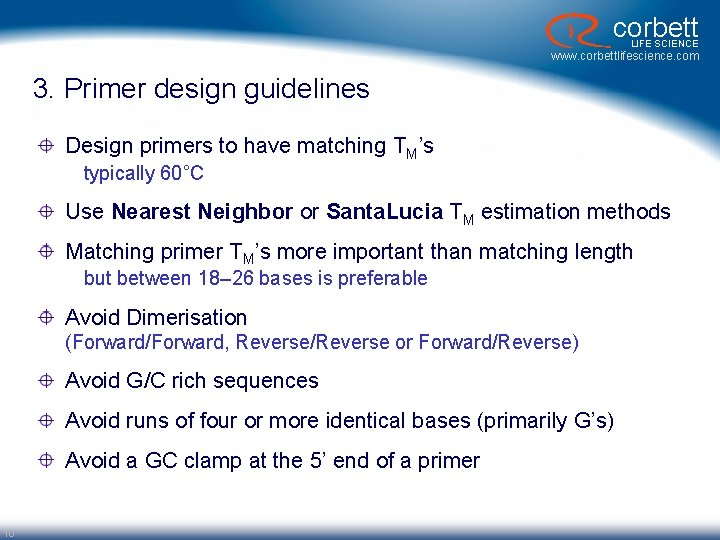
corbett LIFE SCIENCE www. corbettlifescience. com 3. Primer design guidelines Design primers to have matching TM’s typically 60°C Use Nearest Neighbor or Santa. Lucia TM estimation methods Matching primer TM’s more important than matching length but between 18– 26 bases is preferable Avoid Dimerisation (Forward/Forward, Reverse/Reverse or Forward/Reverse) Avoid G/C rich sequences Avoid runs of four or more identical bases (primarily G’s) Avoid a GC clamp at the 5’ end of a primer 10
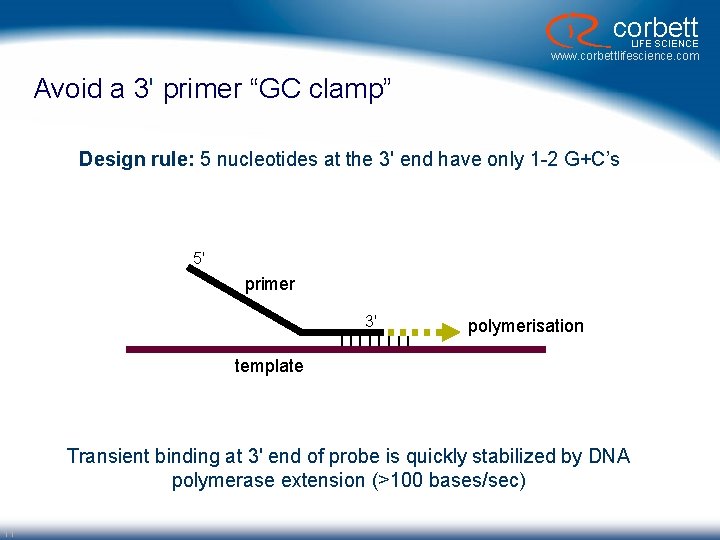
corbett LIFE SCIENCE www. corbettlifescience. com Avoid a 3' primer “GC clamp” Design rule: 5 nucleotides at the 3' end have only 1 -2 G+C’s 5' primer 3' |||| polymerisation template Transient binding at 3' end of probe is quickly stabilized by DNA polymerase extension (>100 bases/sec) 11
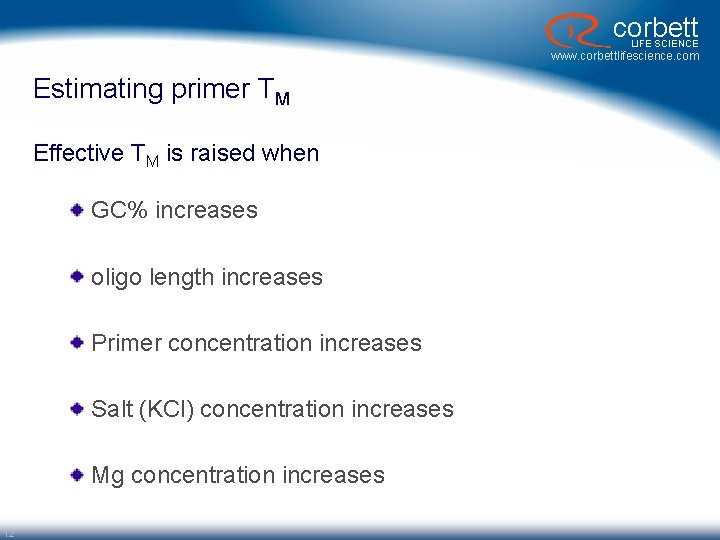
corbett LIFE SCIENCE www. corbettlifescience. com Estimating primer TM Effective TM is raised when GC% increases oligo length increases Primer concentration increases Salt (KCl) concentration increases Mg concentration increases 12
corbett LIFE SCIENCE www. corbettlifescience. com Nearest-Neighbor model Uses empirical data to determine a set of “parameters” for all possible combinations of base pairs. By giving each base-pair a mathematical value, it is possible to quickly and accurately calculate thermodynamics of any hybridization. The nearest neighbor model also accounts for initiation, symmetry, mismatches, dangling ends, internal loops, and bulges. An example shows how the NN model would be used to calculate the d. G of a simple homodimer. Biochemistry. 1996 Mar 19; 35(11): 3555 -62 Improved nearest-neighbor parameters for predicting DNA duplex stability Santa. Lucia J Jr, Allawi HT, Seneviratne PA. Department of Chemistry, Wayne State University, Detroit, Michigan 48202, USA. 13
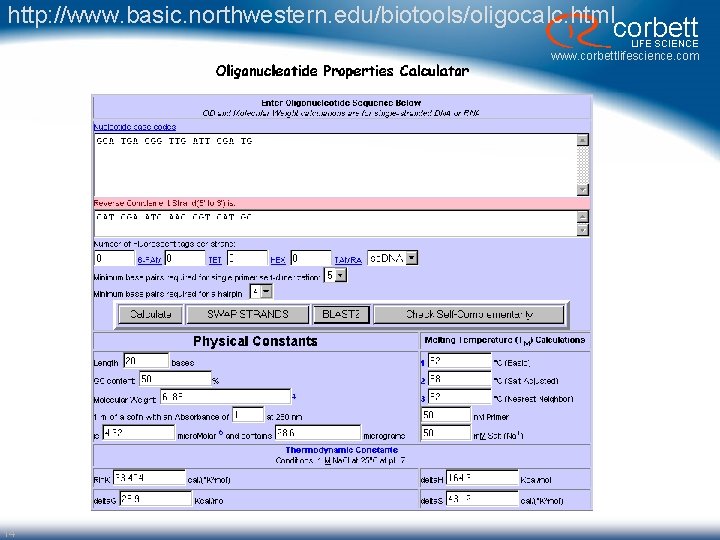
http: //www. basic. northwestern. edu/biotools/oligocalc. html corbett LIFE SCIENCE www. corbettlifescience. com 14
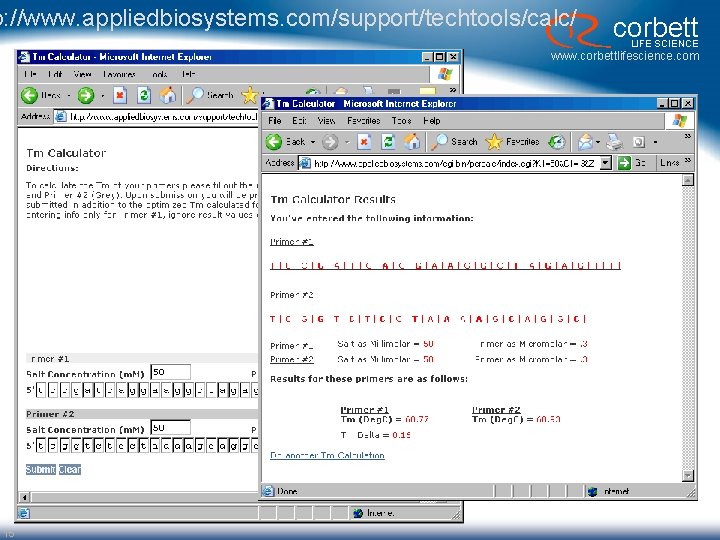
p: //www. appliedbiosystems. com/support/techtools/calc/ corbett LIFE SCIENCE www. corbettlifescience. com 15
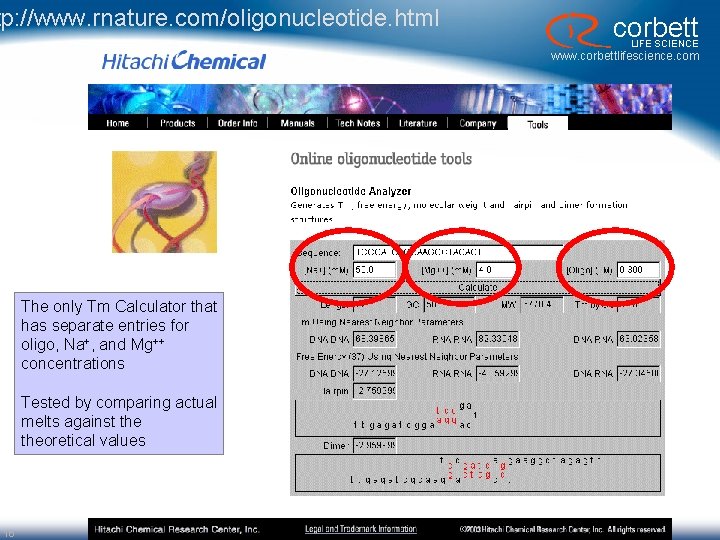
tp: //www. rnature. com/oligonucleotide. html corbett LIFE SCIENCE www. corbettlifescience. com The only Tm Calculator that has separate entries for oligo, Na+, and Mg++ concentrations Tested by comparing actual melts against theoretical values 16
http: //www. biochem. uwo. ca/fac/gloor/oligoform. html corbett LIFE SCIENCE www. corbettlifescience. com University of Western Ontario 17
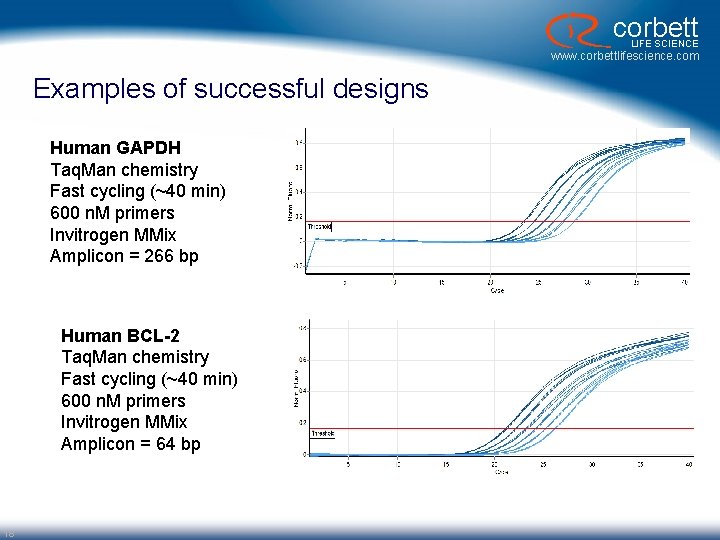
corbett LIFE SCIENCE www. corbettlifescience. com Examples of successful designs Human GAPDH Taq. Man chemistry Fast cycling (~40 min) 600 n. M primers Invitrogen MMix Amplicon = 266 bp Human BCL-2 Taq. Man chemistry Fast cycling (~40 min) 600 n. M primers Invitrogen MMix Amplicon = 64 bp 18
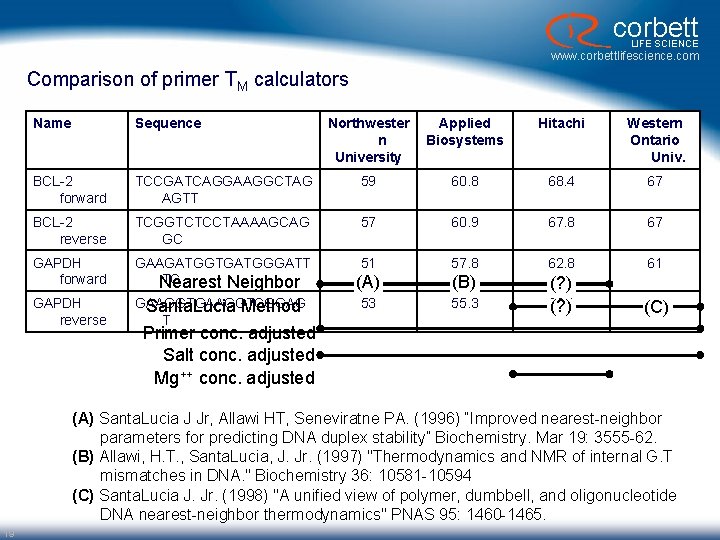
corbett LIFE SCIENCE www. corbettlifescience. com Comparison of primer TM calculators Name Sequence Northwester n University Applied Biosystems Hitachi Western Ontario Univ. BCL-2 forward TCCGATCAGGAAGGCTAG AGTT 59 60. 8 68. 4 67 BCL-2 reverse TCGGTCTCCTAAAAGCAG GC 57 60. 9 67. 8 67 GAPDH forward GAAGATGGTGATGGGATT TC Nearest Neighbor 51 57. 8 62. 8 61 (A) (B) GAPDH reverse GAAGGTCGGAG Santa. Lucia Method T 53 55. 3 (? ) 66. 1 (? ) 65 (C) Primer conc. adjusted Salt conc. adjusted Mg++ conc. adjusted (A) Santa. Lucia J Jr, Allawi HT, Seneviratne PA. (1996) “Improved nearest-neighbor parameters for predicting DNA duplex stability” Biochemistry. Mar 19: 3555 -62. (B) Allawi, H. T. , Santa. Lucia, J. Jr. (1997) "Thermodynamics and NMR of internal G. T mismatches in DNA. " Biochemistry 36: 10581 -10594 (C) Santa. Lucia J. Jr. (1998) "A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics" PNAS 95: 1460 -1465. 19

corbett LIFE SCIENCE www. corbettlifescience. com Tip 1 Design 2 (or 3) forward and reverse primers for each assay Extra primers are cheap (about $1 per base) 2 F + 2 R primers provides 4 different assays 3 F + 3 R primers provides 9 different assays Test all combinations of Forward/Reverse primers to pick best assay: most robust high PCR efficiency works well using high speed cycling 20
corbett LIFE SCIENCE Primer 3 free web software for amplicon design http: //frodo. wi. mit. edu/cgi-bin/primer 3_www. cgi 21 www. corbettlifescience. com

corbett LIFE SCIENCE www. corbettlifescience. com 4. Specific chemistry considerations There are many chemistry types Taq. Man, Eclipse, Beacons, Adjacent dual (FRET) Probes, LUX primers, Scorpion probes, etc There are many chemistry variants MGBs, LNAs, “superbases”, inosine substitutions, etc Trend: the chemistry provider designs the assay Providers understand idiosyncrasies of their chemistry They usually have proprietary design software They usually design for free if you purchase Recommended you always consult chemistry provider about assay design! 22

corbett LIFE SCIENCE www. corbettlifescience. com Example free assay design services MGB Taq. Man Assays-by-design Applied Biosystems LUX primers Invitrogen Beacons, LNAs Proligo MGB Eclipse probes, superbases Qiagen And others… 23

corbett LIFE SCIENCE www. corbettlifescience. com Tip 2 Design a probe for a SYBR assay Allows seamless conversion from a SYBR assay used to screen candidates to a probe assay used for validation 24

corbett LIFE SCIENCE www. corbettlifescience. com Probe: Design Guidelines • Tm: 68 -70 °C for Quantitation • No runs of more than 3 consecutive G’s • No G on 5’end (to avoid quenching of fluorophore) • Select probe from the strand with more C’s than G’s Tm: 65 -67 °C for Allelic Discrimination (SNP Analysis) • Use hot-start Taq polymerase and UNG • Always do a BLAST to check specificity of probe to target 25

corbett LIFE SCIENCE www. corbettlifescience. com What about published assay designs? Don’t assume published assays are optimal Check any existing design for conformity with modern design guidelines Contact the authors before using if possible Web databases of published designs exist 26
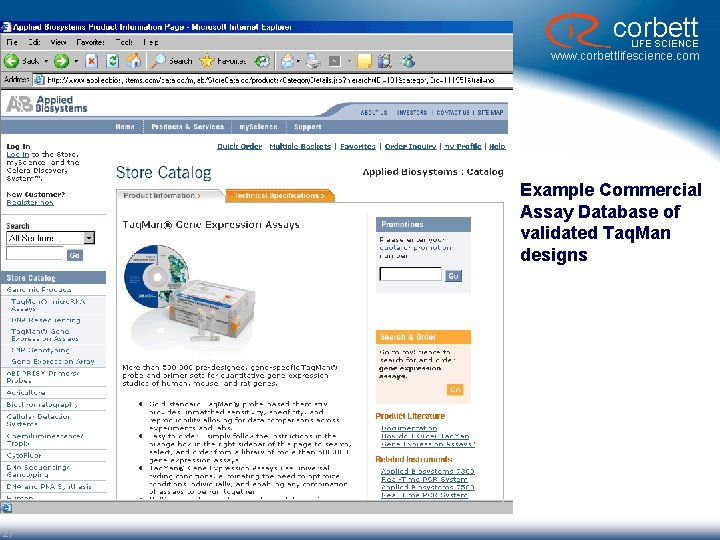
corbett LIFE SCIENCE www. corbettlifescience. com Example Commercial Assay Database of validated Taq. Man designs 27

corbett LIFE SCIENCE www. corbettlifescience. com Example Assay Database http: //www. realtimeprimers. org/ 28
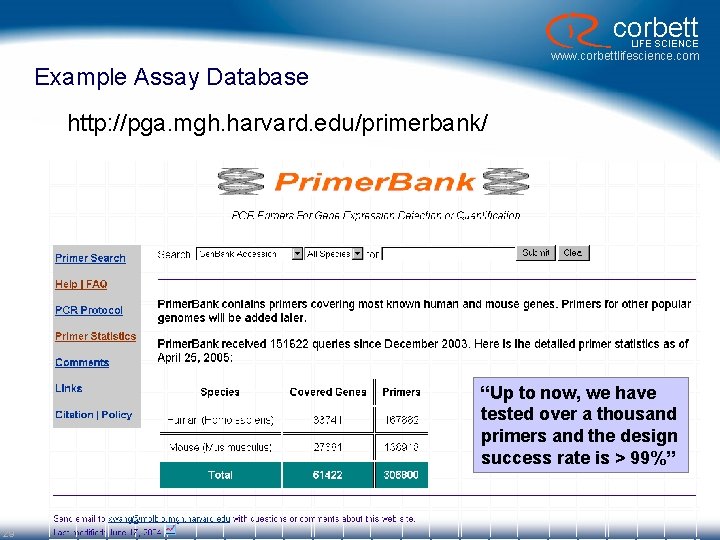
corbett LIFE SCIENCE www. corbettlifescience. com Example Assay Database http: //pga. mgh. harvard. edu/primerbank/ “Up to now, we have tested over a thousand primers and the design success rate is > 99%” 29

corbett LIFE SCIENCE www. corbettlifescience. com Summary Research your target sequence first Avoid the common traps Stick to design guidelines as much as possible They are no guarantee but generally work well Ask for design assistance from chemistry providers Use one TM estimation algorithm They all give different results If you change then match new TMs to your existing assay TMs for consistency Design several primer sets Primer design is key, but success hard to predict Look for pre-existing validated designs Published assays are not always ideal 30

corbett LIFE SCIENCE www. corbettlifescience. com Offices Brisbane Australia Corbett Robotics Pty Ltd 42 Mc. Kechnie Drive Eight Mile Plains, QLD 4113 T +61 7 3841 7077 F +61 7 3841 6077 Sydney Australia Corbett Research Pty Ltd 14 Hilly Street Mortlake, NSW 2137 T +61 2 9736 1320 F +61 2 9736 1364 E-mail info@corbettlifescience. com 32 United Kingdom Corbett Research UK Limited Unit 296 Cambridge Science Park Milton, Cambridge CB 4 0 WD T +44 (0)1223 424 288 F +44 (0)1223 424 144 USA Corbett Robotics Inc China Basin Landing 185 Berry Street, Suite 5200 San Francisco, CA 94107 USA T +1 415 348 1166 F +1 415 348 1177 Web www. corbettlifescience. com All slides 2005 Corbett Life Science. All rights reserved
- Slides: 31