Copy Number Variation Analysis in Gene Panel Sequencing
Copy Number Variation Analysis in Gene Panel Sequencing –Development and validation of an automatic CNV-discovering pipeline at IIHG

Outline for today’s talk 1. Background • The concept of copy number variation and its role in human genetic disease and genetic disease diagnosis. 2. Challenges in CNV analysis and our strategies. 3. Development of automatic CNV-discovering pipeline • Especially the post CNV-call processing for data integration and automatic filtering 4. Results and analysis 5. Summary and path forward
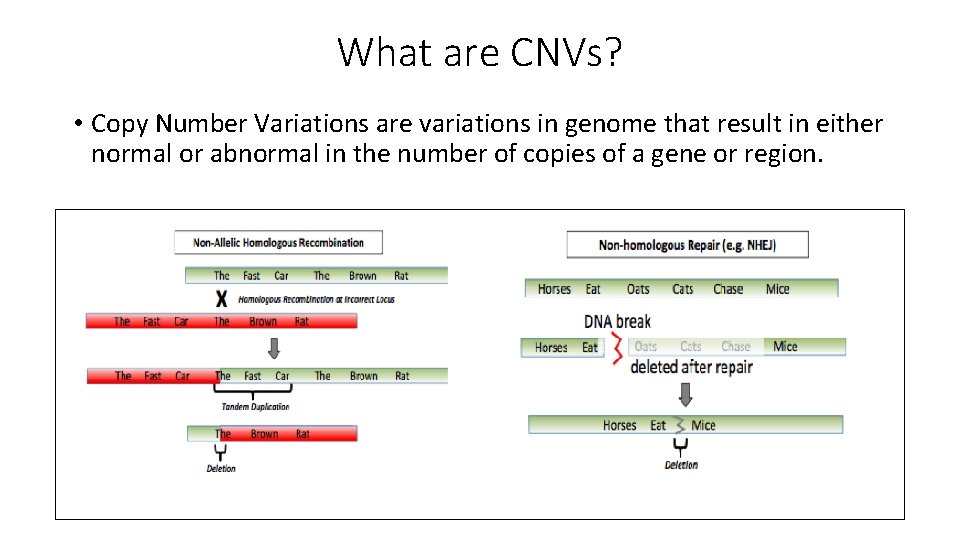
What are CNVs? • Copy Number Variations are variations in genome that result in either normal or abnormal in the number of copies of a gene or region.
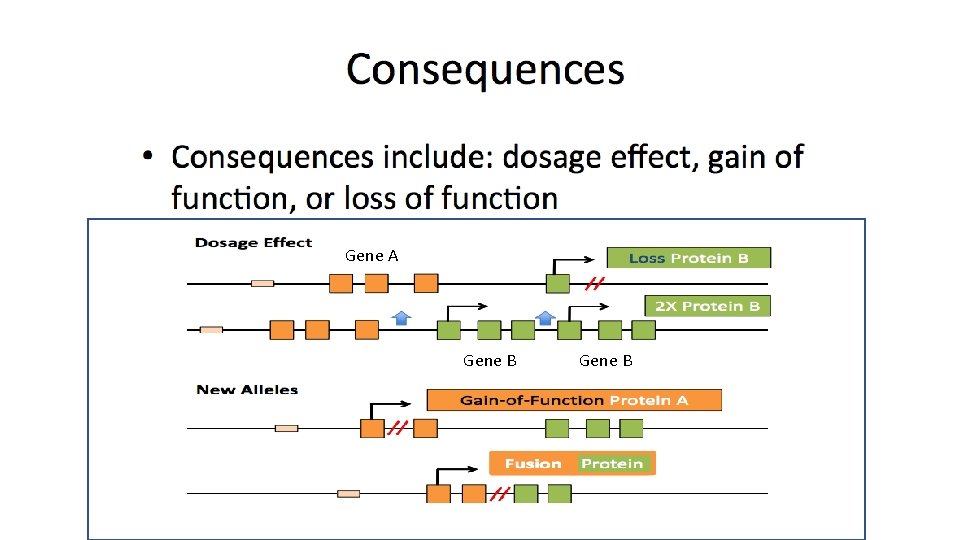
Gene A Gene B

e. g. the deletion of the SHANK 3 gene e. g. CNVs Associated with Susceptibility to Cancers e. g. The deletion variant of DMBT 1 e. g. trisomy 21 e. g. CCL 3 L 1 Copy Number Variation ILD: large genomic imbalances e. g. FCGR 2 B, TNFSF 4, and BANK 1 (losses or gains of e. g. 22 q 11. 2 deletion syndrome. DNA)

CNV analysis, an essential part of our disease diagnosis pipelines • CNV in STRC is one of the most common causes of autosomal recessive non-syndromic hearing loss. • CNV is a common cause of congenital kidney malformations, e. g. deletions at the HNF 1 B locus. • CNVs of CYP 2 D 6, SULT 1 A 1 and UGT 2 B 17 are the major contributors of the metabolism and disposition of many clinically used drugs and xenobiotics. A Eliot Shearer et al. Genome Med. 2014; 6(5): 37. Simone Sanna-Cherchi et al. Am J Hum Genet. 2012 Dec 7; 91(6): 987– 997. Technique report: CYP 2 D 6, SULT 1 A 1 and UGT 2 B 17 copy number variation: quantitative detection by multiplex PCR 2011
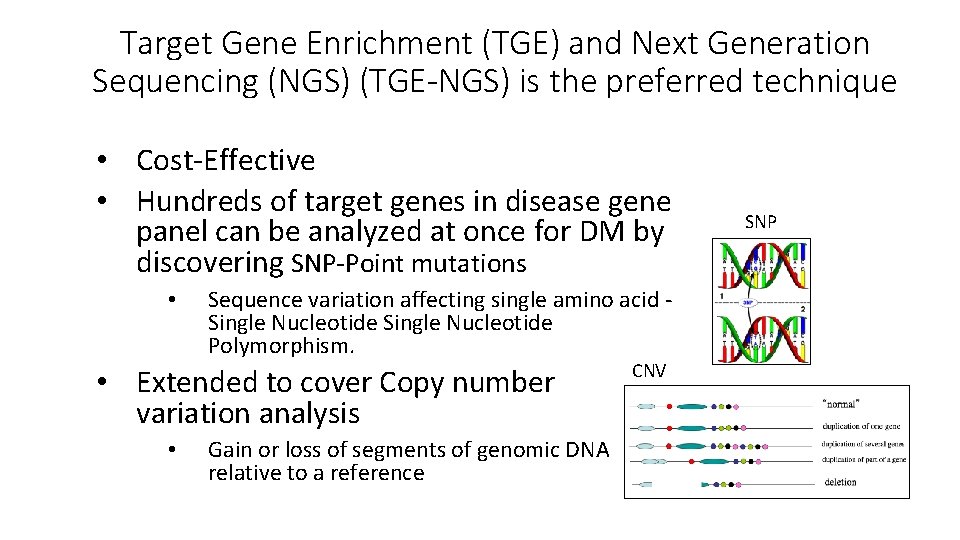
Target Gene Enrichment (TGE) and Next Generation Sequencing (NGS) (TGE-NGS) is the preferred technique • Cost-Effective • Hundreds of target genes in disease gene panel can be analyzed at once for DM by discovering SNP-Point mutations • Sequence variation affecting single amino acid Single Nucleotide Polymorphism. • Extended to cover Copy number variation analysis • Gain or loss of segments of genomic DNA relative to a reference CNV SNP
Challenge in CNV detection from TGE-NGS data • Many exome-based tools available (https: //omictools. com/cnv-detection 3 -category) • Most based on coverage analysis. • Known with extremely high false discover rates. • GC bias: affecting the target affinity, thus hybridization efficiency and the sequence enrichment. • Experimental artifacts.

Challenge in CNV analysis for disease diagnosis • Tens or hundreds of CNV-calls predicted from each run and a majority of them are false positive. • The identification of true positive CNV variants requires manual curation, a step that is laborious, time-consuming, and error-prone.

Goals and Objectives Develop a streamlined CNV-discovering pipeline 1. To reduce false discovery rate, 2. To increase precision and sensitivity (recalls), 3. To automate the CNVdiscovering procedure, 4. To incorporate it as part of IIHG’s standard diagnosis pipelines.

Strategies and Steps taken 1. Evaluated available CNV-discovering tools. 2. Developed strategies for the automatic CNV-discovering procedure (true positive). 3. Implemented as the analysis pipeline with Python as a wrapper 4. Evaluated the pipeline with massive data sets: five batches and a total of 220 patients with hearing loss, additional 100 renal, 150 drug metabolism (DMT) patients using known CNVs as benchmarks: • Manually curated CNVs in patients with hearing loss • Manually curated CNVs in Renal diseases. • CNVs validated by Taq. Man assay in DMT.

Evaluation of publically available CNVdiscovering tools 1. CODEX (Jiang Y, Oldridge DA, Diskin SJ, Zhang NR, Nucleic Acids Res. 2015 Mar 31; 43(6): e 39). Applying Poisson latent factor model to normalize and detect CNVs 2. PANELDOC (Alex S Nord, Ming Lee, Mary-Claire King and Tom Walsh, BMC Genomics 2011 12: 184). Applying PCA or other statistic approaches to normalize and detect CNVs 3. Exome. Copy (Love MI, Myšičková A, Sun R, Kalscheuer V, Vingron M, Haas SA, Stat Appl Genet Mol Biol. 2011 Nov 8; 10(1). ) Applying negative binomial distributions to normalize and detect CNVs 4. Exome. Depth (Vincent Plagnol, James Curtis et al. Bioinformatics (2012) 28 (21): 2747 -2754). Using a group of samples as a control 5. cnv. Kit (Eric Talevich, A. Hunter Shain, Thomas Botton, Boris C. Bastian, 2016 PLo. S Comput Biol 12(4): e 1004873). Using a group of samples as a control

Lessons and knowledge learned from the tool evaluation 1. Differ greatly in ability for CNV identifications • Exomedepth and cnv. Kit with the highest precision • Panel. Do. C and Exome. Copy with the highest sensitivity • Codex with a limited ability in CNV discovery. 2. Their ability also differ among the type of genes and diseases (hearing loss, renal and DMT). 3. All tools have high false discovery rates, Panel. Do. C and Exome. Copy are the highest. 4. All tools have specific statistic parameters to measure the confidence in CNV-discovery but CNV identified with the strongest signals are not necessarily true positive.
Strategies: a method of multiple tool assembling Hypothesis: • Persistent Signaling features of true positive CNV (relative to those resulted from random experimental artifacts. • True positive CNV would be discovered with a high probability no matter what CNV-discovering tools are applied. Observation: The majority of the false positive calls were found to be attributable to different normalization and discovery algorithms, therefore, unique to a particular caller (Hong et. al, 2016). Concordance analysis among CNV callers: Allow CNV-analysis with reduced false positive rates and increased precision and sensitivity. T 2 T 1 T 3
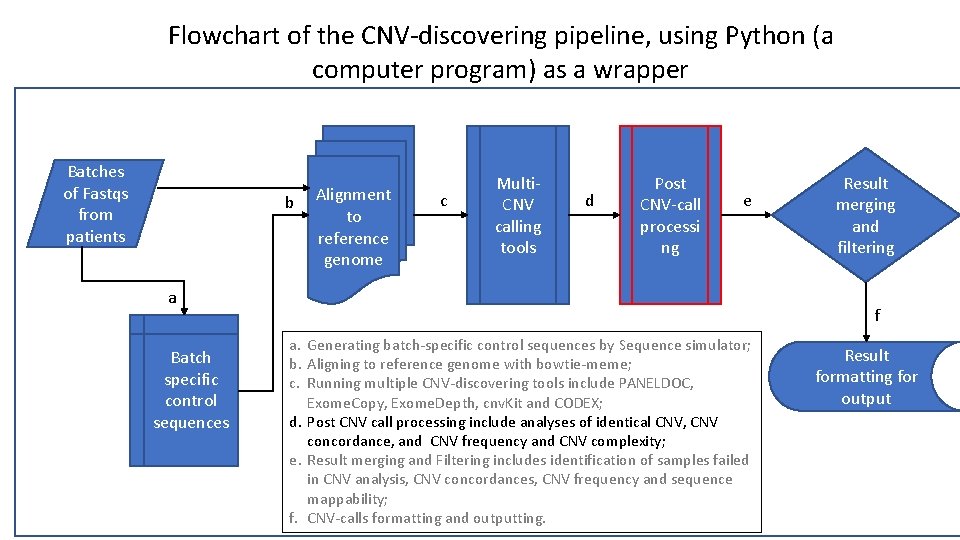
Flowchart of the CNV-discovering pipeline, using Python (a computer program) as a wrapper Batches of Fastqs from patients b Alignment to reference genome c Multi. CNV calling tools d Post CNV-call processi ng e a Batch specific control sequences Result merging and filtering f a. Generating batch-specific control sequences by Sequence simulator; b. Aligning to reference genome with bowtie-meme; c. Running multiple CNV-discovering tools include PANELDOC, Exome. Copy, Exome. Depth, cnv. Kit and CODEX; d. Post CNV call processing include analyses of identical CNV, CNV concordance, and CNV frequency and CNV complexity; e. Result merging and Filtering includes identification of samples failed in CNV analysis, CNV concordances, CNV frequency and sequence mappability; f. CNV-calls formatting and outputting. Result formatting for output
Post CNV-call processing for data integration and automatic filtering Defined several key concepts associated with merging, filtering and formatting, include a. The CNV complexity (for sample filtering) b. Identical (equivalent) CNV calls. c. cnv. Freq (CNV frequency). d. S-Scoring (concordance scoring).
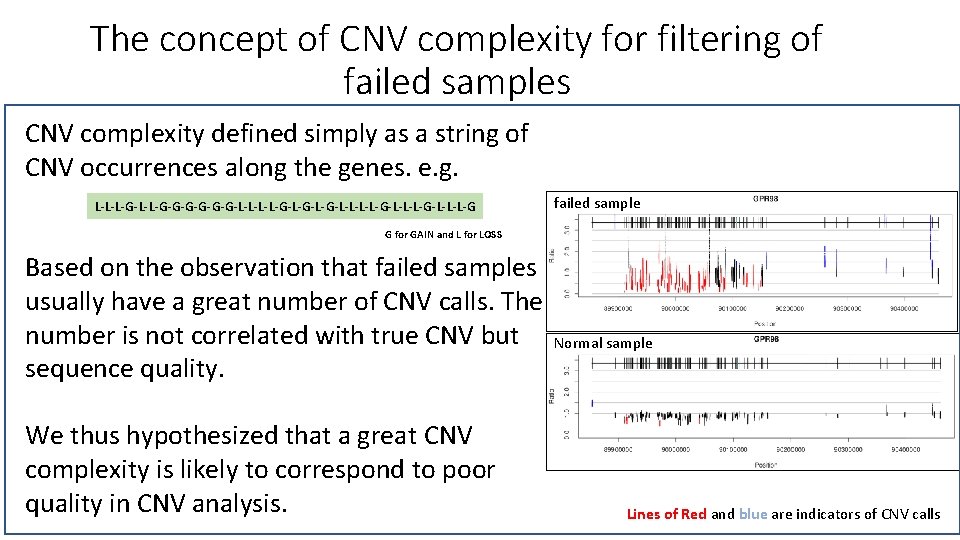
The concept of CNV complexity for filtering of failed samples CNV complexity defined simply as a string of CNV occurrences along the genes. e. g. L-L-L-G-G-G-G-L-L-G-L-G-L-L-G-L-L-L-G failed sample G for GAIN and L for LOSS Based on the observation that failed samples usually have a great number of CNV calls. The number is not correlated with true CNV but sequence quality. We thus hypothesized that a great CNV complexity is likely to correspond to poor quality in CNV analysis. Normal sample Lines of Red and blue are indicators of CNV calls
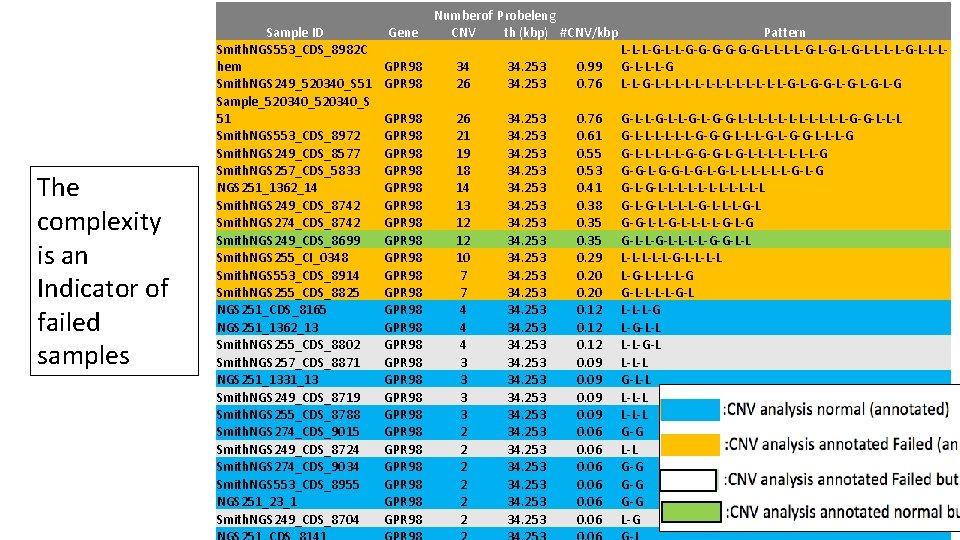
The complexity is an Indicator of failed samples Sample ID Smith. NGS 553_CDS_8982 C hem Smith. NGS 249_520340_S 51 Sample_520340_S 51 Smith. NGS 553_CDS_8972 Smith. NGS 249_CDS_8577 Smith. NGS 257_CDS_5833 NGS 251_1362_14 Smith. NGS 249_CDS_8742 Smith. NGS 274_CDS_8742 Smith. NGS 249_CDS_8699 Smith. NGS 255_CI_0348 Smith. NGS 553_CDS_8914 Smith. NGS 255_CDS_8825 NGS 251_CDS_8165 NGS 251_1362_13 Smith. NGS 255_CDS_8802 Smith. NGS 257_CDS_8871 NGS 251_1331_13 Smith. NGS 249_CDS_8719 Smith. NGS 255_CDS_8788 Smith. NGS 274_CDS_9015 Smith. NGS 249_CDS_8724 Smith. NGS 274_CDS_9034 Smith. NGS 553_CDS_8955 NGS 251_23_1 Smith. NGS 249_CDS_8704 Gene Numberof Probeleng CNV th (kbp) #CNV/kbp GPR 98 34 26 34. 253 0. 99 0. 76 Pattern L-L-L-G-G-G-G-L-L-G-L-G-L-L-G-L-L-L-G-L-L-L-L-L-L-L-G-G-L-G-L-G GPR 98 GPR 98 GPR 98 GPR 98 GPR 98 GPR 98 26 21 19 18 14 13 12 12 10 7 7 4 4 4 3 3 2 2 2 34. 253 34. 253 34. 253 0. 76 0. 61 0. 55 0. 53 0. 41 0. 38 0. 35 0. 29 0. 20 0. 12 0. 09 0. 06 G-L-L-G-L-G-G-L-L-L-L-L-L-G-G-L-L-L-G-G-G-L-L-L-G-G-L-L-L-G-G-G-L-L-L-L-G G-G-L-G-L-L-L-L-G-L-G G-L-L-L-L-L-L-L G-L-L-L-L-G-L G-G-L-L-L-L-G-G-L-L L-L-L-G-L-L-G G-L-L-G-L L-L-L-G L-G-L-L L-L-G-L L-L-L G-L-L L-L-L Orange G-G Yellow L-L G-G Green G-G Blue L-G
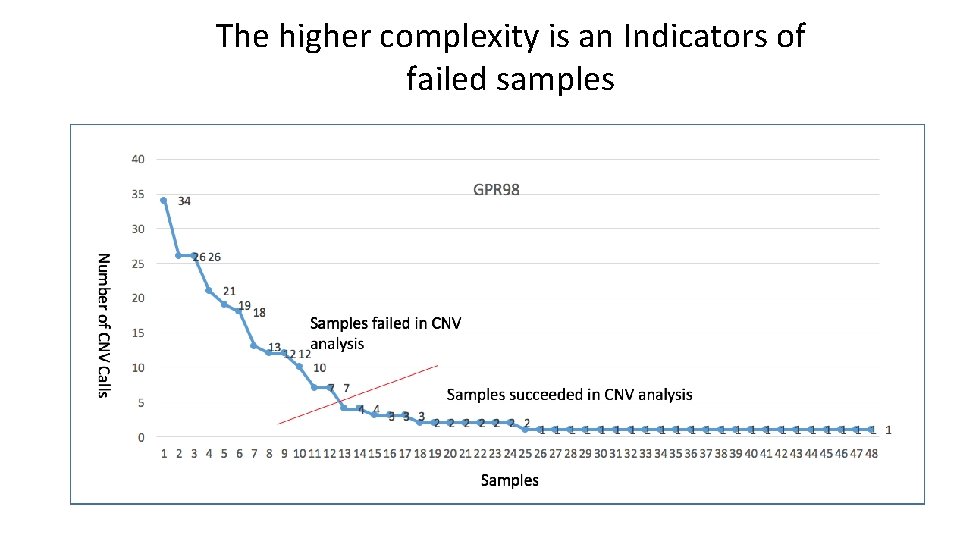
The higher complexity is an Indicators of failed samples
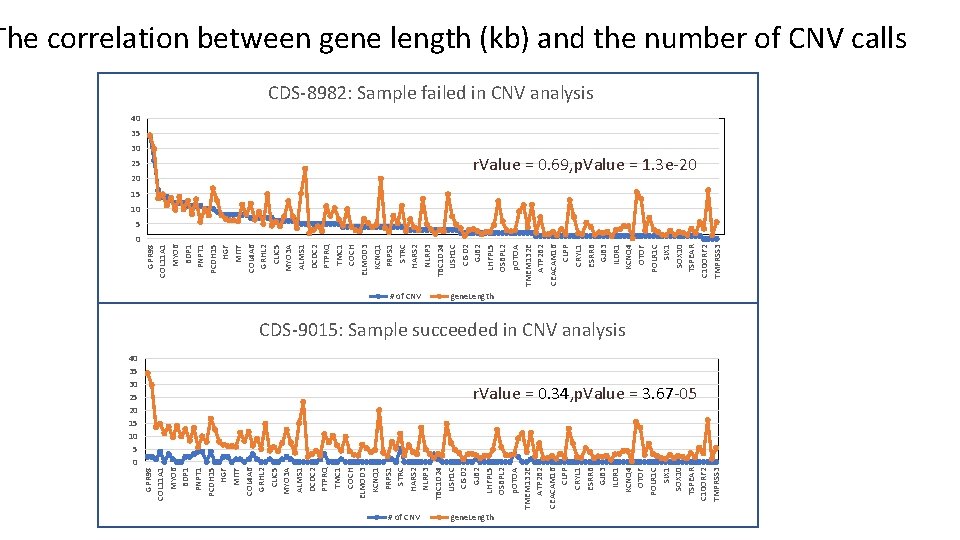
40 35 30 25 20 15 10 5 0 GPR 98 COL 11 A 1 MYO 6 BDP 1 PNPT 1 PCDH 15 HGF MITF COL 4 A 6 GRHL 2 CLIC 5 MYO 3 A ALMS 1 DCDC 2 PTPRQ TMC 1 COCH ELMOD 3 KCNQ 1 PRPS 1 STRC HARS 2 NLRP 3 TBC 1 D 24 USH 1 C CISD 2 GJB 2 LHFPL 5 OSBPL 2 p. OTOA TMEM 132 E ATP 2 B 2 CEACAM 16 CLPP CRYL 1 ESRRB GJB 3 ILDR 1 KCNQ 4 OTOF POLR 1 C SIX 1 SOX 10 TSPEAR C 10 ORF 2 TMPRSS 3 The correlation between gene length (kb) and the number of CNV calls CDS-8982: Sample failed in CNV analysis 40 35 30 25 20 r. Value = 0. 69, p. Value = 1. 3 e-20 15 10 5 # of CNV gene. Length CDS-9015: Sample succeeded in CNV analysis r. Value = 0. 34, p. Value = 3. 67 -05 gene. Length
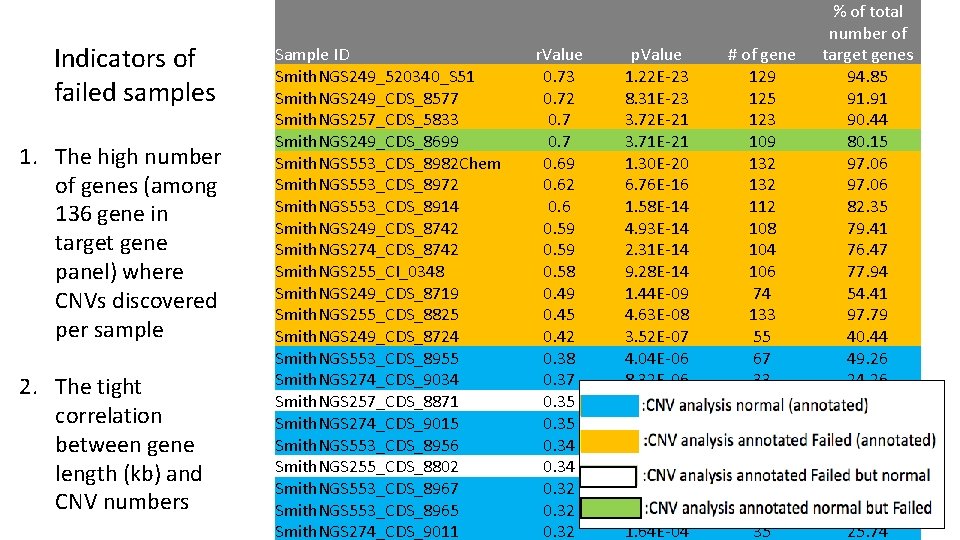
Indicators of failed samples 1. The high number of genes (among 136 gene in target gene panel) where CNVs discovered per sample 2. The tight correlation between gene length (kb) and CNV numbers Sample ID Smith. NGS 249_520340_S 51 Smith. NGS 249_CDS_8577 Smith. NGS 257_CDS_5833 Smith. NGS 249_CDS_8699 Smith. NGS 553_CDS_8982 Chem Smith. NGS 553_CDS_8972 Smith. NGS 553_CDS_8914 Smith. NGS 249_CDS_8742 Smith. NGS 274_CDS_8742 Smith. NGS 255_CI_0348 Smith. NGS 249_CDS_8719 Smith. NGS 255_CDS_8825 Smith. NGS 249_CDS_8724 Smith. NGS 553_CDS_8955 Smith. NGS 274_CDS_9034 Smith. NGS 257_CDS_8871 Smith. NGS 274_CDS_9015 Smith. NGS 553_CDS_8956 Smith. NGS 255_CDS_8802 Smith. NGS 553_CDS_8967 Smith. NGS 553_CDS_8965 Smith. NGS 274_CDS_9011 r. Value p. Value 0. 73 1. 22 E-23 0. 72 8. 31 E-23 0. 7 3. 72 E-21 0. 7 3. 71 E-21 0. 69 1. 30 E-20 0. 62 6. 76 E-16 0. 6 1. 58 E-14 0. 59 4. 93 E-14 0. 59 2. 31 E-14 0. 58 9. 28 E-14 0. 49 1. 44 E-09 0. 45 4. 63 E-08 0. 42 3. 52 E-07 0. 38 4. 04 E-06 0. 37 8. 32 E-06 0. 35 Orange 2. 88 E-05 0. 35 3. 67 E-05 0. 34 Yellow 5. 89 E-05 0. 34 Green 4. 90 E-05 0. 32 1. 37 E-04 0. 32 Blue 1. 26 E-04 0. 32 1. 64 E-04 # of gene 129 125 123 109 132 112 108 104 106 74 133 55 67 33 23 35 74 48 36 39 35 % of total number of target genes 94. 85 91. 91 90. 44 80. 15 97. 06 82. 35 79. 41 76. 47 77. 94 54. 41 97. 79 40. 44 49. 26 24. 26 16. 91 25. 74 54. 41 35. 29 26. 47 28. 68 25. 74
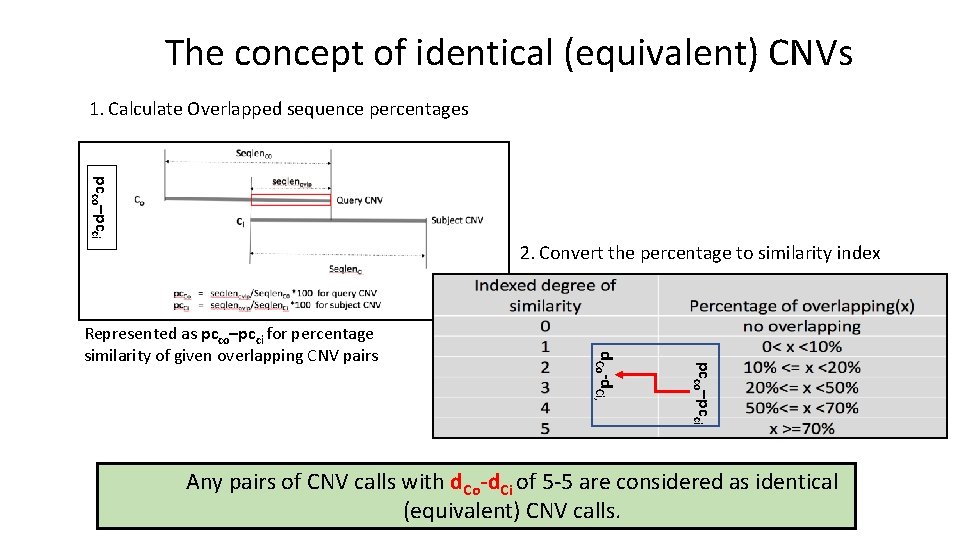
The concept of identical (equivalent) CNVs 1. Calculate Overlapped sequence percentages pcco–pcci 2. Convert the percentage to similarity index pcco–pcci d. Co-d. Ci, Represented as pcco–pcci for percentage similarity of given overlapping CNV pairs Any pairs of CNV calls with d. Co-d. Ci of 5 -5 are considered as identical (equivalent) CNV calls.

cnv. Freq for automatic CNV filtering Observations: Frequency of disease-causing CNV mutation is often quite rare, except STRC/OTOA in hearing losses, and CYP 2 D 6 in drug metabolisms. Hypotheses: A high frequency of predicted CNV calls may be due to platform-specific artifacts (experiment procedures), or common CNV variants. A CNV frequency would provide a good filtering to enrich positive CNV calls.
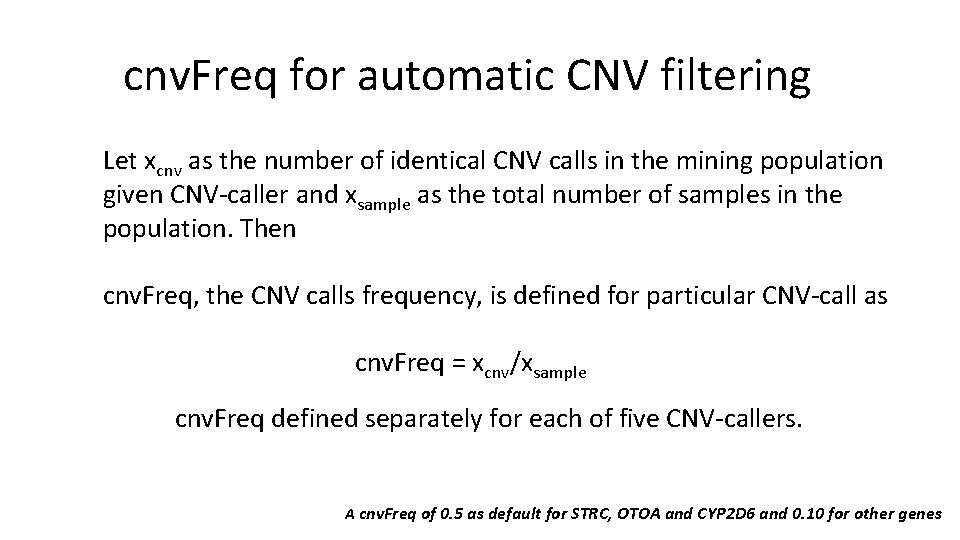
cnv. Freq for automatic CNV filtering Let xcnv as the number of identical CNV calls in the mining population given CNV-caller and xsample as the total number of samples in the population. Then cnv. Freq, the CNV calls frequency, is defined for particular CNV-call as cnv. Freq = xcnv/xsample cnv. Freq defined separately for each of five CNV-callers. A cnv. Freq of 0. 5 as default for STRC, OTOA and CYP 2 D 6 and 0. 10 for other genes
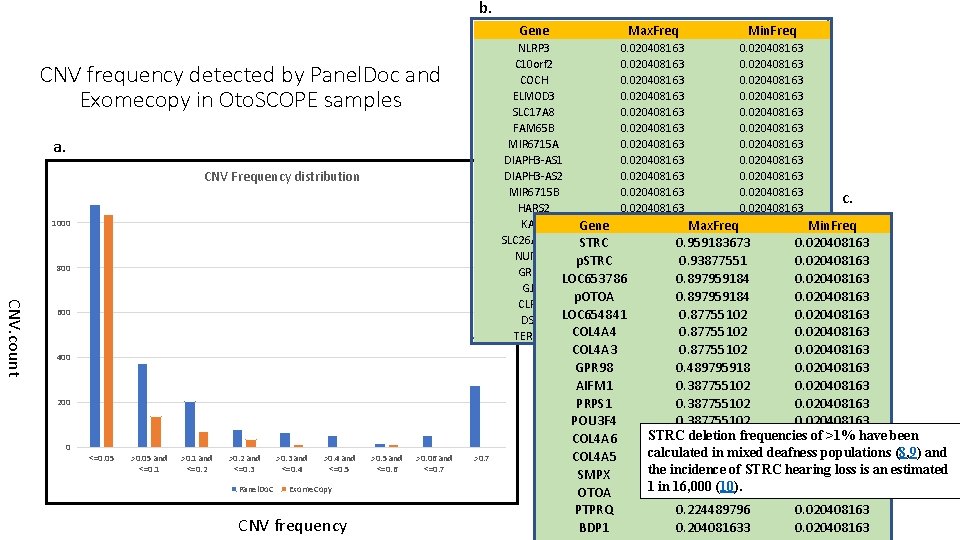
b. Gene a. CNV Frequency distribution 1000 800 CNV. count 600 400 200 <=0. 05 >0. 05 and <=0. 1 >0. 1 and <=0. 2 >0. 2 and <=0. 3 Panel. Do. C >0. 3 and <=0. 4 >0. 4 and <=0. 5 Exome. Copy CNV frequency >0. 5 and <=0. 6 >0. 06 and <=0. 7 Min. Freq NLRP 3 0. 020408163 C 10 orf 2 0. 020408163 COCH 0. 020408163 ELMOD 3 0. 020408163 SLC 17 A 8 0. 020408163 FAM 65 B 0. 020408163 MIR 6715 A 0. 020408163 DIAPH 3 -AS 1 0. 020408163 DIAPH 3 -AS 2 0. 020408163 MIR 6715 B 0. 020408163 c. HARS 2 0. 020408163 KARS Gene 0. 020408163 Max. Freq 0. 020408163 Min. Freq SLC 26 A 4 -AS 1 STRC 0. 020408163 0. 959183673 0. 020408163 NUMA 1 0. 020408163 p. STRC 0. 93877551 0. 020408163 GRHL 2 0. 020408163 LOC 653786 0. 897959184 0. 020408163 GJB 6 0. 020408163 p. OTOA 0. 897959184 0. 020408163 CLRN 1 0. 020408163 DSPP LOC 6548410. 0204081630. 87755102 0. 020408163 COL 4 A 4 0. 0204081630. 877551020. 020408163 TERF 2 IP CNV frequency detected by Panel. Doc and Exomecopy in Oto. SCOPE samples 0 Max. Freq >0. 7 COL 4 A 3 GPR 98 AIFM 1 PRPS 1 POU 3 F 4 COL 4 A 6 COL 4 A 5 SMPX OTOA PTPRQ BDP 1 0. 87755102 0. 020408163 0. 489795918 0. 020408163 0. 387755102 0. 020408163 STRC deletion frequencies of >1% have been 0. 367346939 0. 020408163 calculated in mixed deafness populations (8, 9) and 0. 367346939 0. 020408163 the incidence of STRC hearing loss is an estimated 0. 367346939 0. 020408163 1 in 16, 000 (10). 0. 265306122 0. 020408163 0. 224489796 0. 020408163 0. 204081633 0. 020408163
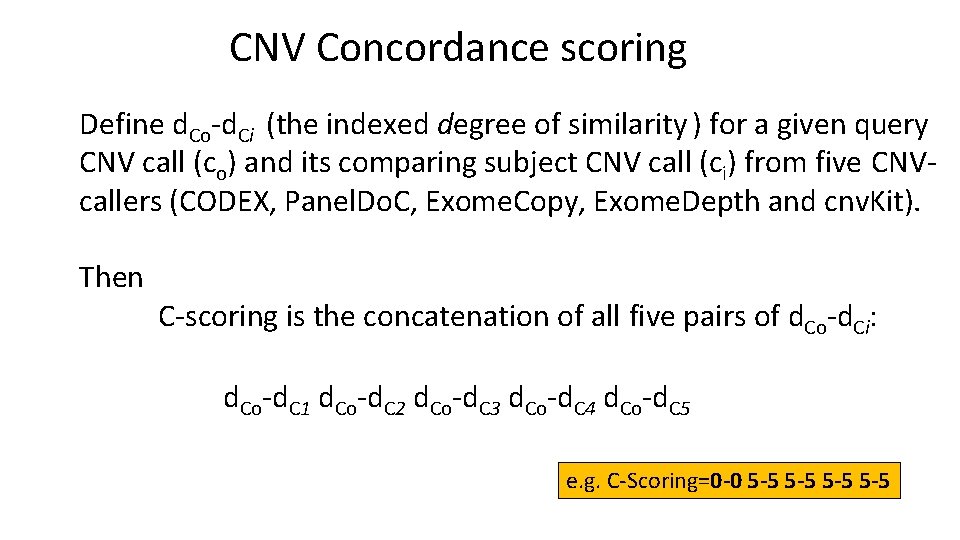
CNV Concordance scoring Define d. Co-d. Ci (the indexed degree of similarity ) for a given query CNV call (co) and its comparing subject CNV call (ci) from five CNVcallers (CODEX, Panel. Do. C, Exome. Copy, Exome. Depth and cnv. Kit). Then C-scoring is the concatenation of all five pairs of d. Co-d. Ci: d. Co-d. C 1 d. Co-d. C 2 d. Co-d. C 3 d. Co-d. C 4 d. Co-d. C 5 e. g. C-Scoring=0 -0 5 -5 5 -5
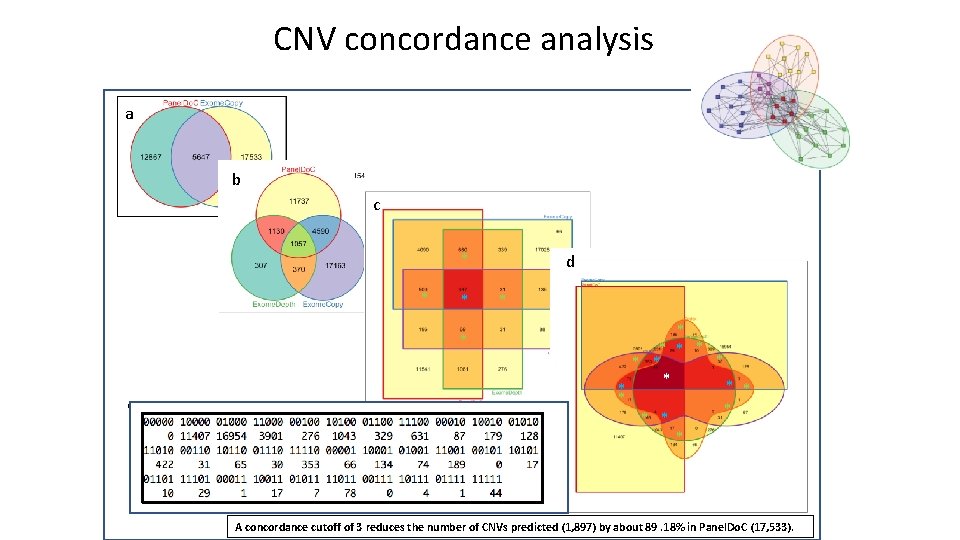
CNV concordance analysis a b c * * e d * * * * * A concordance cutoff of 3 reduces the number of CNVs predicted (1, 897) by about 89. 18% in Panel. Do. C (17, 533).
An efficient transition from manual to automation in CNV-discovering Manual Hard labor Time-consuming Error-pruning il f V N at m to am s ic d. C n a e l p ng i r te Automation u le ib s s o ep a by d Ma High precision and sensitivity
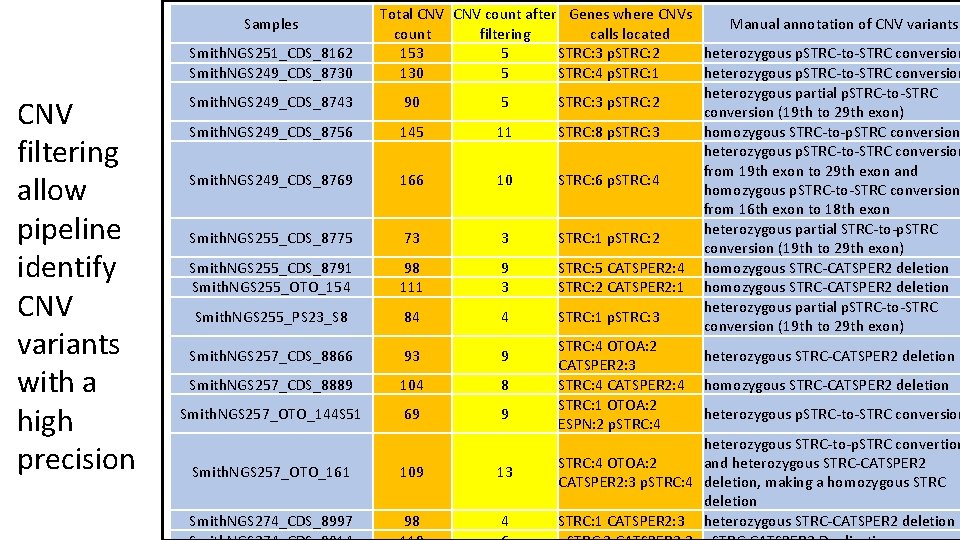
Samples Smith. NGS 251_CDS_8162 Smith. NGS 249_CDS_8730 CNV filtering allow pipeline identify CNV variants with a high precision Smith. NGS 249_CDS_8743 Smith. NGS 249_CDS_8756 Smith. NGS 249_CDS_8769 Smith. NGS 255_CDS_8775 Smith. NGS 255_CDS_8791 Smith. NGS 255_OTO_154 Smith. NGS 255_PS 23_S 8 Smith. NGS 257_CDS_8866 Smith. NGS 257_CDS_8889 Smith. NGS 257_OTO_144 S 51 Smith. NGS 257_OTO_161 Smith. NGS 274_CDS_8997 Total CNV count after Genes where CNVs Manual annotation of CNV variants count filtering calls located 153 5 STRC: 3 p. STRC: 2 heterozygous p. STRC-to-STRC conversion 130 5 STRC: 4 p. STRC: 1 heterozygous p. STRC-to-STRC conversion heterozygous partial p. STRC-to-STRC 90 5 STRC: 3 p. STRC: 2 conversion (19 th to 29 th exon) 145 11 STRC: 8 p. STRC: 3 homozygous STRC-to-p. STRC conversion heterozygous p. STRC-to-STRC conversion from 19 th exon to 29 th exon and 166 10 STRC: 6 p. STRC: 4 homozygous p. STRC-to-STRC conversion from 16 th exon to 18 th exon heterozygous partial STRC-to-p. STRC 73 3 STRC: 1 p. STRC: 2 conversion (19 th to 29 th exon) 98 9 STRC: 5 CATSPER 2: 4 homozygous STRC-CATSPER 2 deletion 111 3 STRC: 2 CATSPER 2: 1 homozygous STRC-CATSPER 2 deletion heterozygous partial p. STRC-to-STRC 84 4 STRC: 1 p. STRC: 3 conversion (19 th to 29 th exon) STRC: 4 OTOA: 2 93 9 heterozygous STRC-CATSPER 2 deletion CATSPER 2: 3 104 8 STRC: 4 CATSPER 2: 4 homozygous STRC-CATSPER 2 deletion STRC: 1 OTOA: 2 69 9 heterozygous p. STRC-to-STRC conversion ESPN: 2 p. STRC: 4 heterozygous STRC-to-p. STRC convertion STRC: 4 OTOA: 2 and heterozygous STRC-CATSPER 2 109 13 CATSPER 2: 3 p. STRC: 4 deletion, making a homozygous STRC deletion 98 4 STRC: 1 CATSPER 2: 3 heterozygous STRC-CATSPER 2 deletion
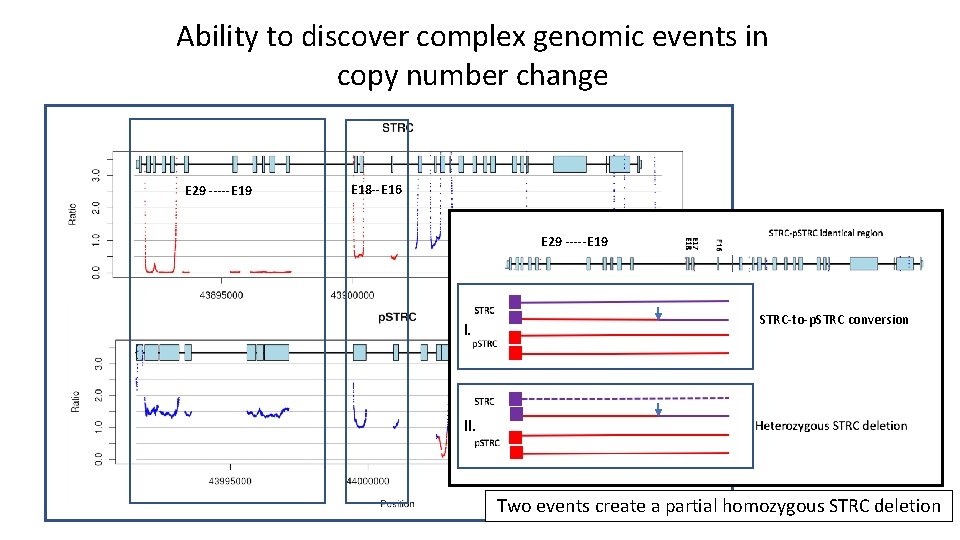
Ability to discover complex genomic events in copy number change E 29 -----E 19 E 18 --E 16 E 29 -----E 19 I. STRC-to-p. STRC conversion II. Two events create a partial homozygous STRC deletion
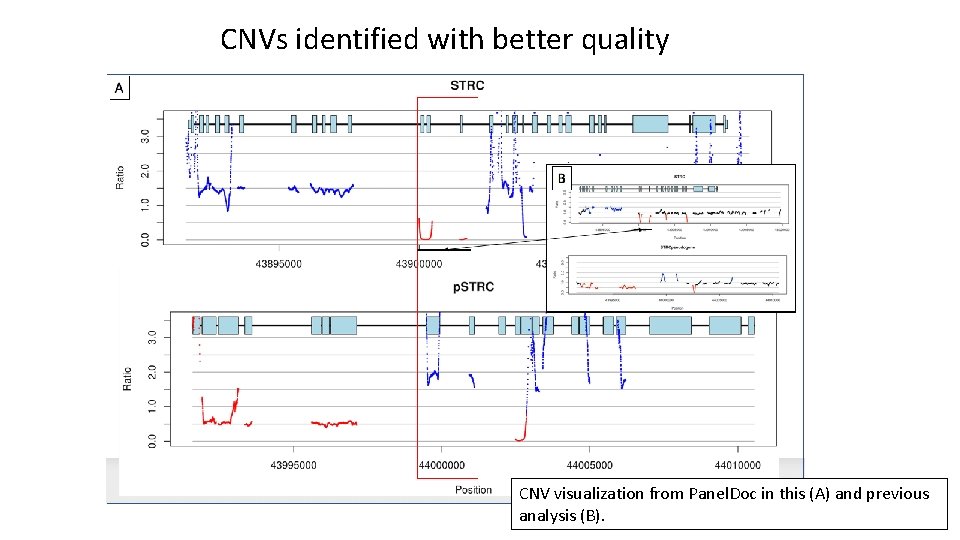
CNVs identified with better quality CNV visualization from Panel. Doc in this (A) and previous analysis (B).
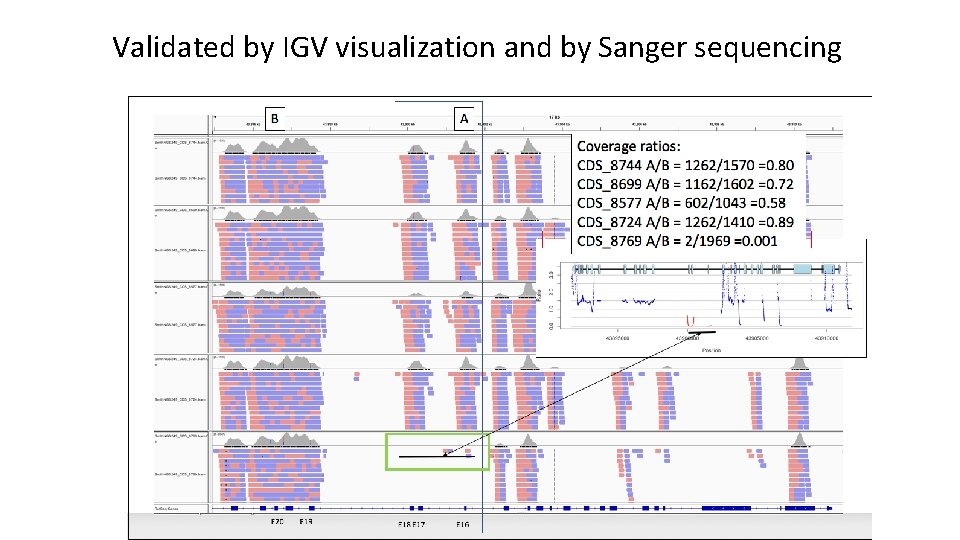
Validated by IGV visualization and by Sanger sequencing
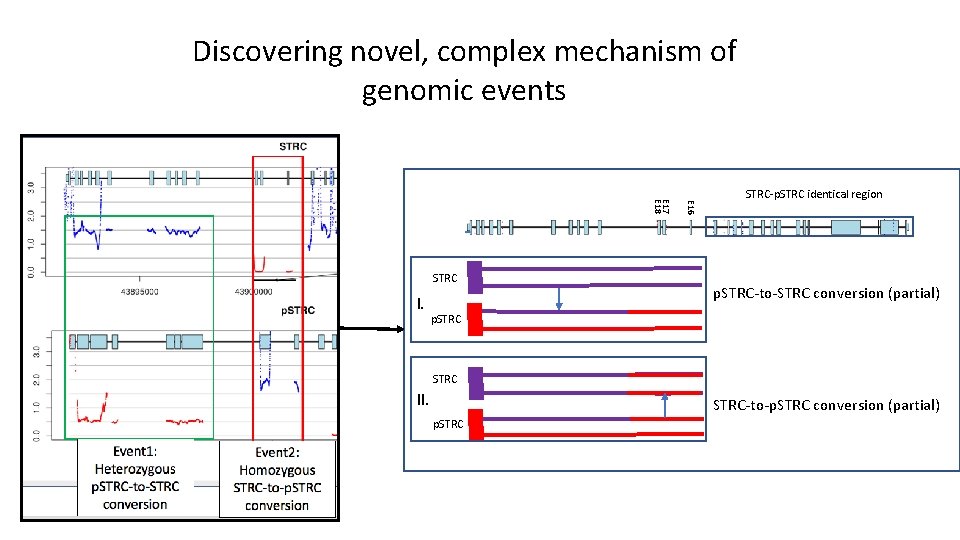
Discovering novel, complex mechanism of genomic events I. E 16 E 17 E 18 STRC-p. STRC identical region p. STRC-to-STRC conversion (partial) p. STRC II. STRC-to-p. STRC conversion (partial) p. STRC
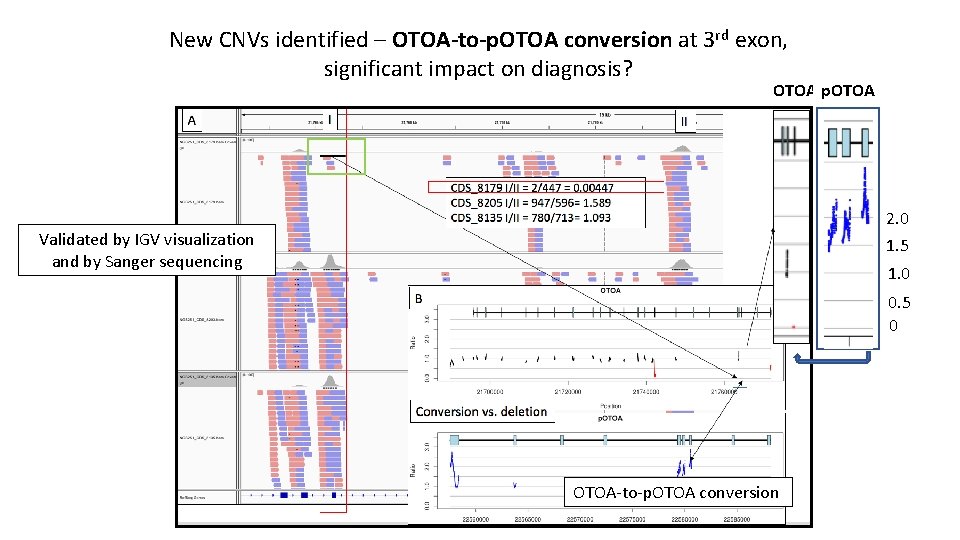
New CNVs identified – OTOA-to-p. OTOA conversion at 3 rd exon, significant impact on diagnosis? OTOA p. OTOA 2. 0 1. 5 Validated by IGV visualization and by Sanger sequencing 1. 0 0. 5 0 OTOA-to-p. OTOA conversion
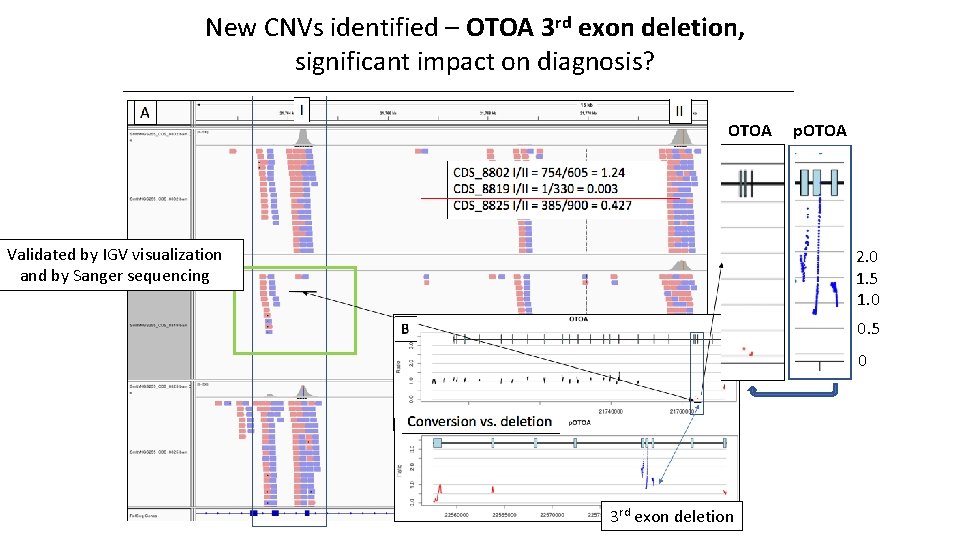
New CNVs identified – OTOA 3 rd exon deletion, significant impact on diagnosis? OTOA Validated by IGV visualization and by Sanger sequencing p. OTOA 2. 0 1. 5 1. 0 0. 5 0 3 rd exon deletion
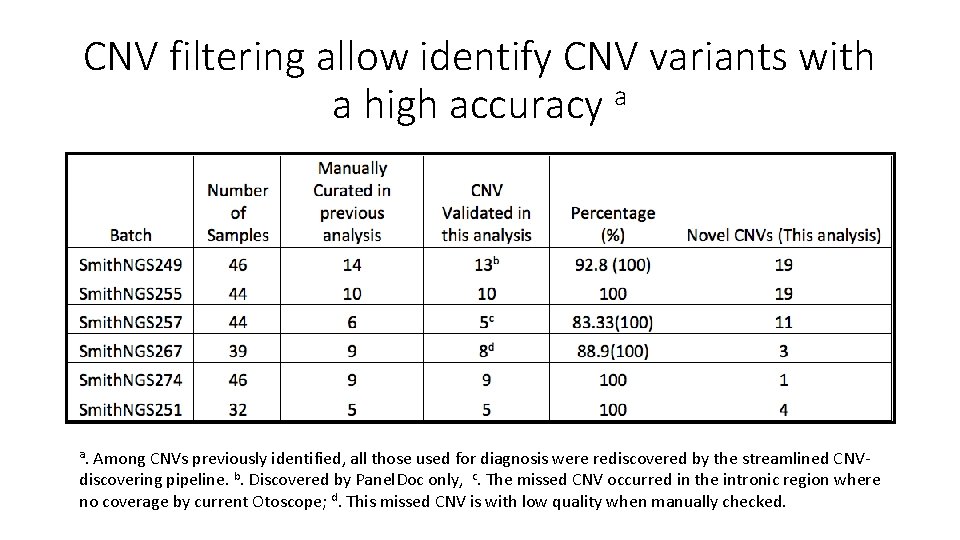
CNV filtering allow identify CNV variants with a high accuracy a a. Among CNVs previously identified, all those used for diagnosis were rediscovered by the streamlined CNVdiscovering pipeline. b. Discovered by Panel. Doc only, c. The missed CNV occurred in the intronic region where no coverage by current Otoscope; d. This missed CNV is with low quality when manually checked.
Summary and path forward 1. Developed a prototype of CNV analysis pipeline with assembling of 5 different CNV callers. 2. Implemented efficient strategies for CNV merging and filtering. 3. Evaluated the pipeline with total of 220 patients with hearing loss, additional 100 Renal and 150 DMT patients. a. Significantly reduced number of false discovered CNVs with increased precision and sensitivity. b. Validated the majority of CNVs from previous manual analysis. All those causative CNVs previously identified were rediscovered. c. New CNVs discovered that potentially change the diagnosis results. 4. More work planned for additional testing, validation and eventual integration.

Acknowledgement Sompallae, Ramakrishna Chimenti, Michael Crone, Bradley Wertz, Julie S Azaiez, Hela Wang, Donghong (Maggie) Kashmola, Iman E Koizumi, Lisa Kimble, Mycah Kolbe, Diana L Bair, Thomas B Nishimura, Carla Campbell, Colleen Kwitek, Anne E Smith, Richard J

Thanks and Questions

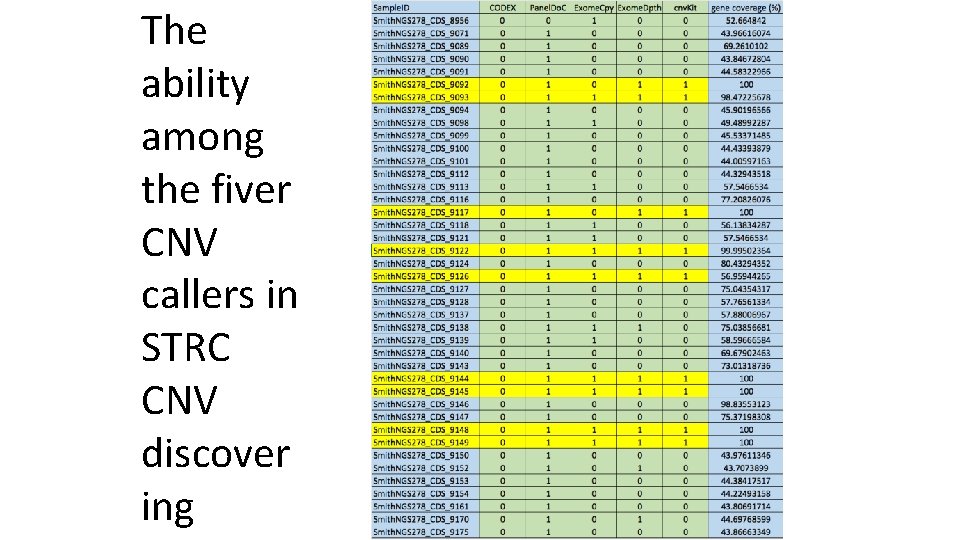
The ability among the fiver CNV callers in STRC CNV discover ing
- Slides: 41