Construct a Highdensity Genetic Map and Its Application






- Slides: 45

Construct a High-density Genetic Map and Its Application to Genetic Regulation Analysis for Yield and Fiber Quality Traits Youlu Yuan Institute of Cotton Research, CAAS State Key Laboratory of Cotton Biology China yuanyoulu@caas. cn

♦ Cotton plays a important role in the world economy by producing natural fiber for textile industry and edible oil for human consumption ♦ Modern high-speed textile operations around the world require long, strong and fine cotton fibers.

♦Conventional breeding procedure had difficulty in further improving fiber quality due to its high cost, long duration, and low selective efficiency. ♦ DNA markers provide an opportunity to overcome the shortcoming of conventional breeding in cotton.

The progress of cotton genome research in China the sequencing of (Gossypium raimondii)(2012 , 2013) the sequencing of Gossypium arboreum)(2014) the sequencing of Gossypium hirsutum and Gossypium barbadense(2015, 2016) 1. Nature Genetics, 2012 2. Nature , 2012 3. Nature Gen, 2014 4. Nature Biotech, 2015 5. Nature biotech, 2015 6. Scientific Reports, 2015 7. Scientific Reports, 2015

Major marker u. Genetics map construction with SSR markers u. Genetics map construction with SNP markers discovered by cotton 63 K chip u. Genetics map construction with SNP markers discovered by SLAF-Seq (specific locus amplified fragment sequencing ) u. Genome-wide consensus map construction and its utilization

Genetics map construction and QTL identification with SSR markers

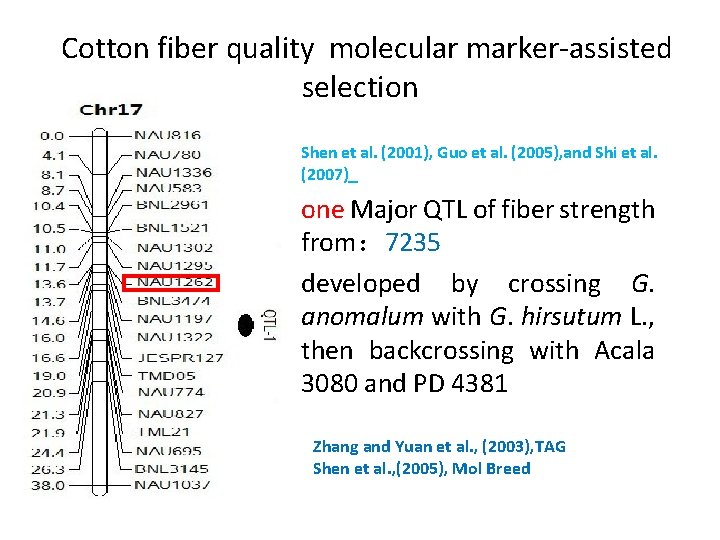
Cotton fiber quality molecular marker-assisted selection Shen et al. (2001), Guo et al. (2005), and Shi et al. (2007) one Major QTL of fiber strength from: 7235 developed by crossing G. anomalum with G. hirsutum L. , then backcrossing with Acala 3080 and PD 4381 Zhang and Yuan et al. , (2003), TAG Shen et al. , (2005), Mol Breed

Up to 2001, A few stable or common QTLs have been reported ►►Therefore, we constructed F 2, F 2: 3, and RILs populations to screen stable QTL associated to fiber quality across multiple generations and environments.

Parents selection 0 -153: G. arboreum introgression line with superior fiber quality from Sichuan agricultural university s. GK 9708: the selected line from the commercial transgenic cultivar CCRI 41, resistant to budworm, bred in cotton research institute, CAAS, China s. GK 9708

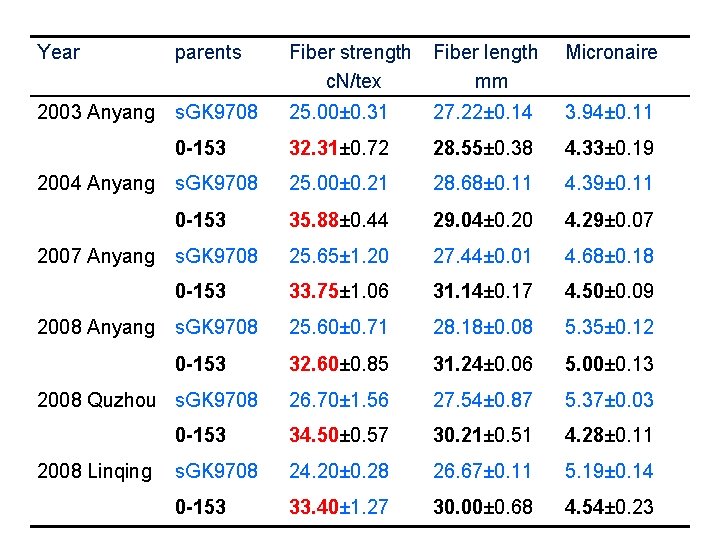
Year parents Fiber strength c. N/tex Fiber length mm Micronaire 2003 Anyang s. GK 9708 25. 00± 0. 31 27. 22± 0. 14 3. 94± 0. 11 0 -153 32. 31± 0. 72 28. 55± 0. 38 4. 33± 0. 19 s. GK 9708 25. 00± 0. 21 28. 68± 0. 11 4. 39± 0. 11 0 -153 35. 88± 0. 44 29. 04± 0. 20 4. 29± 0. 07 s. GK 9708 25. 65± 1. 20 27. 44± 0. 01 4. 68± 0. 18 0 -153 33. 75± 1. 06 31. 14± 0. 17 4. 50± 0. 09 s. GK 9708 25. 60± 0. 71 28. 18± 0. 08 5. 35± 0. 12 0 -153 32. 60± 0. 85 31. 24± 0. 06 5. 00± 0. 13 26. 70± 1. 56 27. 54± 0. 87 5. 37± 0. 03 0 -153 34. 50± 0. 57 30. 21± 0. 51 4. 28± 0. 11 s. GK 9708 24. 20± 0. 28 26. 67± 0. 11 5. 19± 0. 14 0 -153 33. 40± 1. 27 30. 00± 0. 68 4. 54± 0. 23 2004 Anyang 2007 Anyang 2008 Quzhou s. GK 9708 2008 Linqing

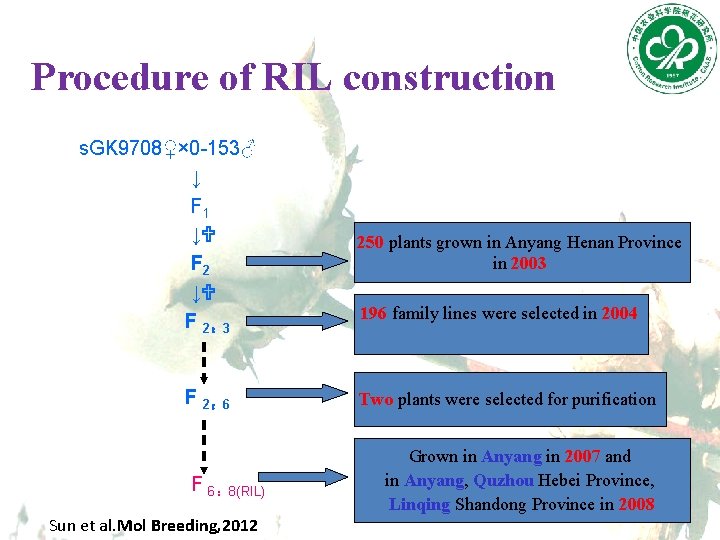
Procedure of RIL construction s. GK 9708♀× 0 -153♂ ↓ F 1 ↓U F 2: 3 F 2: 6 F 6: 8(RIL) Sun et al. Mol Breeding, 2012 250 plants grown in Anyang Henan Province in 2003 196 family lines were selected in 2004 Two plants were selected for purification Grown in Anyang in 2007 and in Anyang, Quzhou Hebei Province, Linqing Shandong Province in 2008

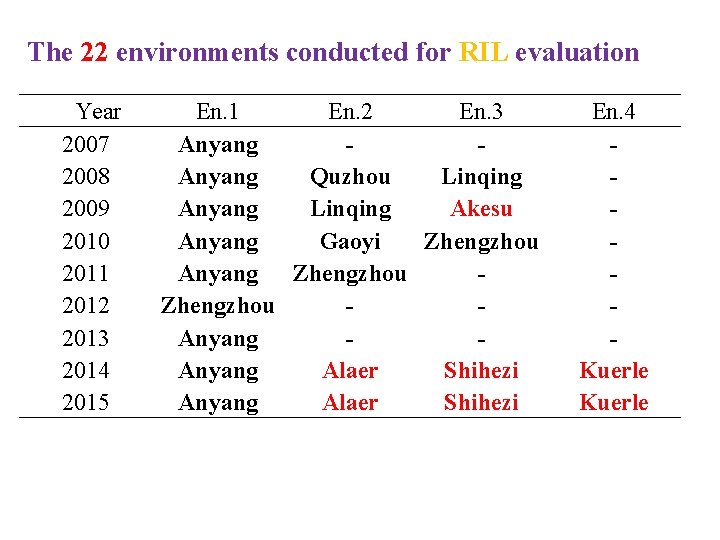
The 22 environments conducted for RIL evaluation Year 2007 2008 2009 2010 2011 2012 2013 2014 2015 En. 1 En. 2 En. 3 Anyang Quzhou Linqing Anyang Linqing Akesu Anyang Gaoyi Zhengzhou Anyang Alaer Shihezi Anyang Alaer Shihezi En. 4 Kuerle

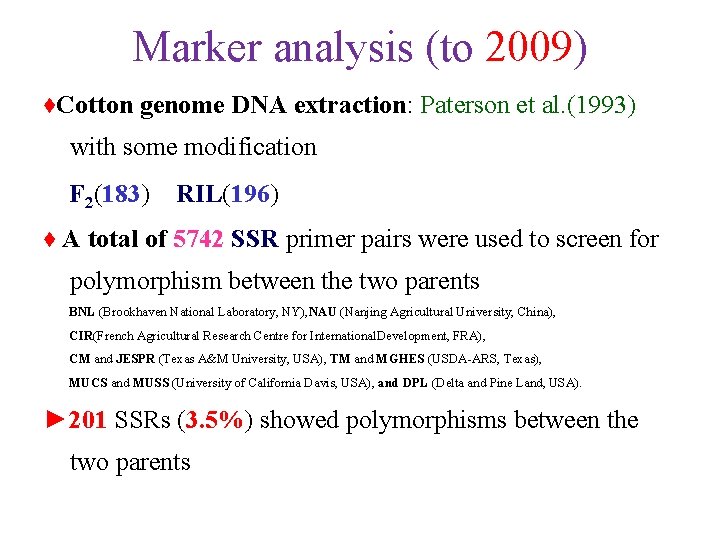
Marker analysis (to 2009) ♦Cotton genome DNA extraction: Paterson et al. (1993) with some modification F 2(183) RIL(196) ♦ A total of 5742 SSR primer pairs were used to screen for polymorphism between the two parents BNL (Brookhaven National Laboratory, NY), NAU (Nanjing Agricultural University, China), CIR(French Agricultural Research Centre for International. Development, FRA), CM and JESPR (Texas A&M University, USA), TM and MGHES (USDA-ARS, Texas), MUCS and MUSS (University of California Davis, USA), and DPL (Delta and Pine Land, USA). ► 201 SSRs (3. 5%) showed polymorphisms between the two parents

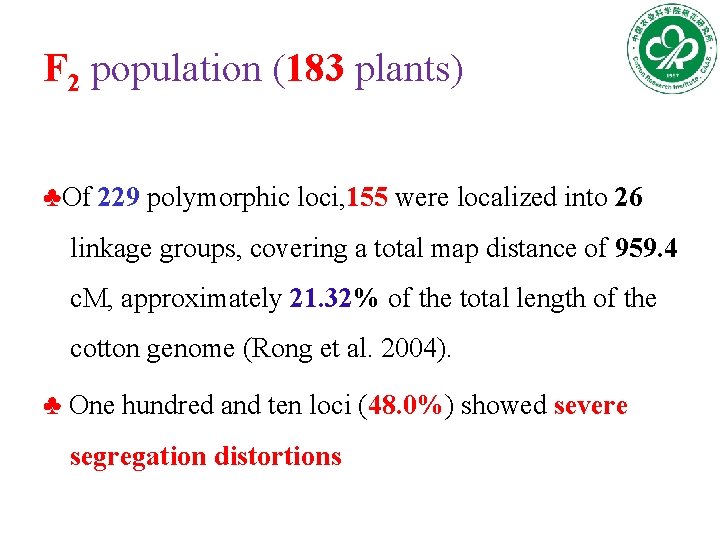
F 2 population (183 plants) ♣Of 229 polymorphic loci, 155 were localized into 26 linkage groups, covering a total map distance of 959. 4 c. M, approximately 21. 32% of the total length of the cotton genome (Rong et al. 2004). ♣ One hundred and ten loci (48. 0%) showed severe segregation distortions

Part of linkage groups for F 2 population

RIL population (196 lines) ♣ Of 217 polymorphic loci, 190 loci were localized into 34 linkage groups with a total map distance of 700. 9 c. M, approximately 15. 75% of the total length of the cotton genome. ♣ Severe segregation distortions were observed with 93 (43. 3%) out of 217 loci.

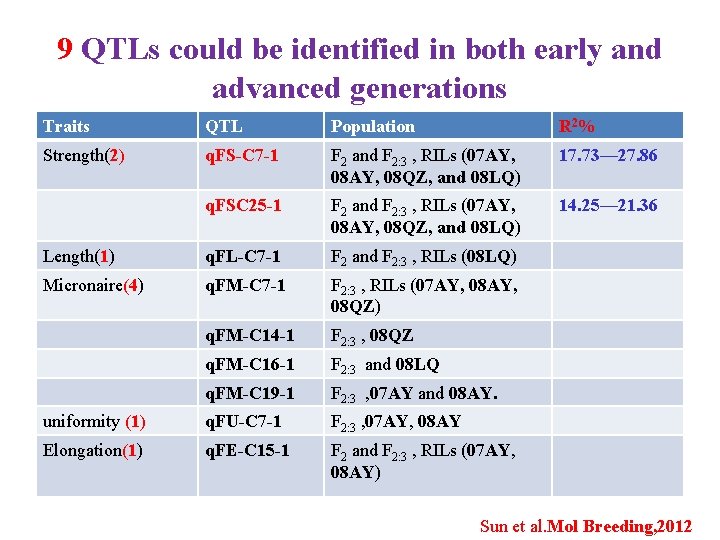
9 QTLs could be identified in both early and advanced generations Traits QTL Population R 2 % Strength(2) q. FS-C 7 -1 F 2 and F 2: 3 , RILs (07 AY, 08 QZ, and 08 LQ) 17. 73— 27. 86 q. FSC 25 -1 F 2 and F 2: 3 , RILs (07 AY, 08 QZ, and 08 LQ) 14. 25— 21. 36 Length(1) q. FL-C 7 -1 F 2 and F 2: 3 , RILs (08 LQ) Micronaire(4) q. FM-C 7 -1 F 2: 3 , RILs (07 AY, 08 QZ) q. FM-C 14 -1 F 2: 3 , 08 QZ q. FM-C 16 -1 F 2: 3 and 08 LQ q. FM-C 19 -1 F 2: 3 , 07 AY and 08 AY. uniformity (1) q. FU-C 7 -1 F 2: 3 , 07 AY, 08 AY Elongation(1) q. FE-C 15 -1 F 2 and F 2: 3 , RILs (07 AY, 08 AY) Sun et al. Mol Breeding, 2012

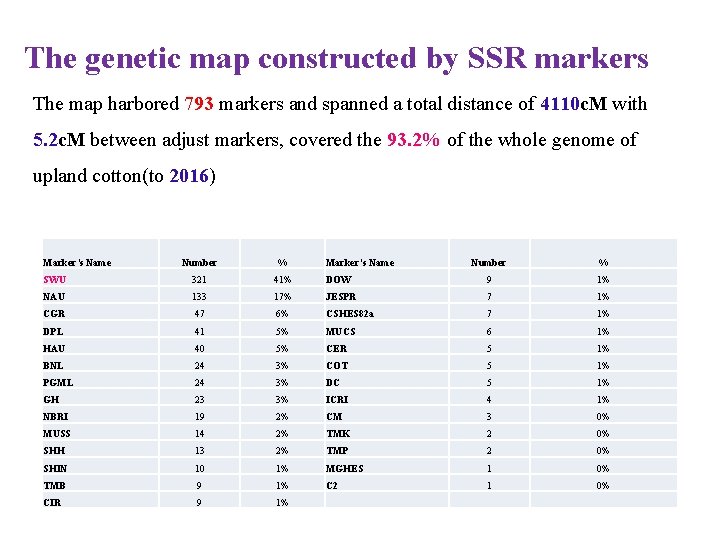
The genetic map constructed by SSR markers The map harbored 793 markers and spanned a total distance of 4110 c. M with 5. 2 c. M between adjust markers, covered the 93. 2% of the whole genome of upland cotton(to 2016) Marker’s Name Number % SWU 321 41% NAU 133 CGR Marker’s Name Number % DOW 9 1% 17% JESPR 7 1% 47 6% CSHES 82 a 7 1% DPL 41 5% MUCS 6 1% HAU 40 5% CER 5 1% BNL 24 3% COT 5 1% PGML 24 3% DC 5 1% GH 23 3% ICRI 4 1% NBRI 19 2% CM 3 0% MUSS 14 2% TMK 2 0% SHH 13 2% TMP 2 0% SHIN 10 1% MGHES 1 0% TMB 9 1% C 2 1 0% CIR 9 1%

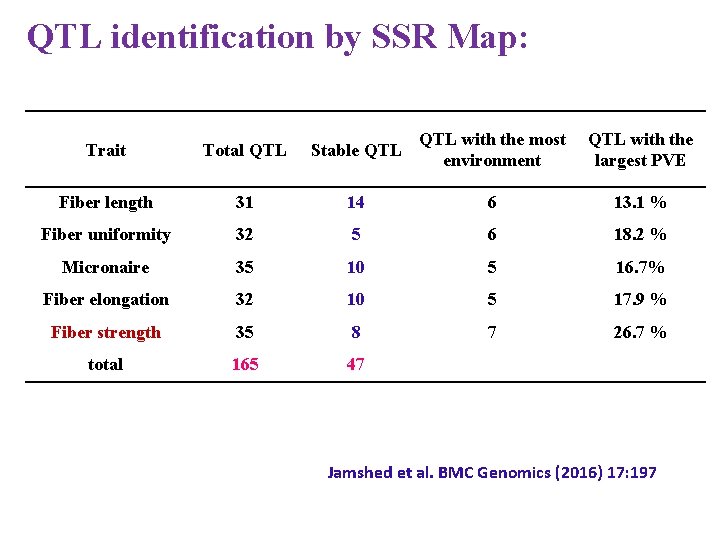
QTL identification by SSR Map: Trait Total QTL Stable QTL with the most environment QTL with the largest PVE Fiber length 31 14 6 13. 1 % Fiber uniformity 32 5 6 18. 2 % Micronaire 35 10 5 16. 7% Fiber elongation 32 10 5 17. 9 % Fiber strength 35 8 7 26. 7 % total 165 47 Jamshed et al. BMC Genomics (2016) 17: 197

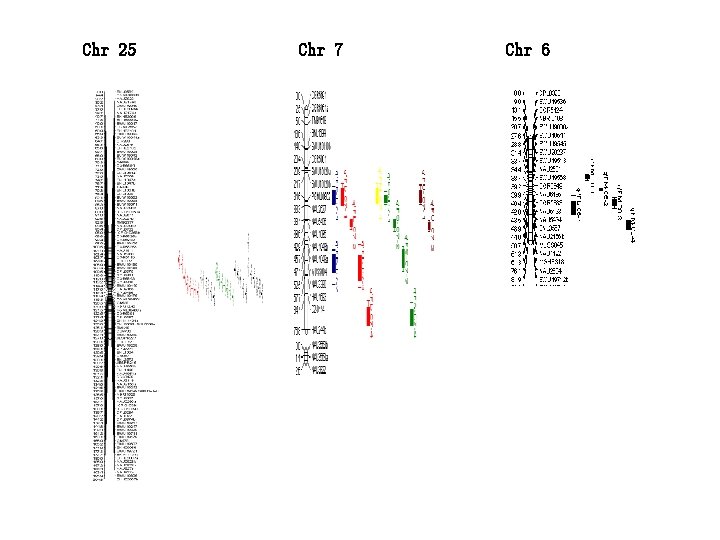
Chr 25 Chr 7 Chr 6

Genetics map construction and QTL identification with SNP markers discovered by cotton 63 K chip Zhang zhen etal. , Crop Sci. 2017,57: 774– 788

Map constructed by 63 K SNP Array

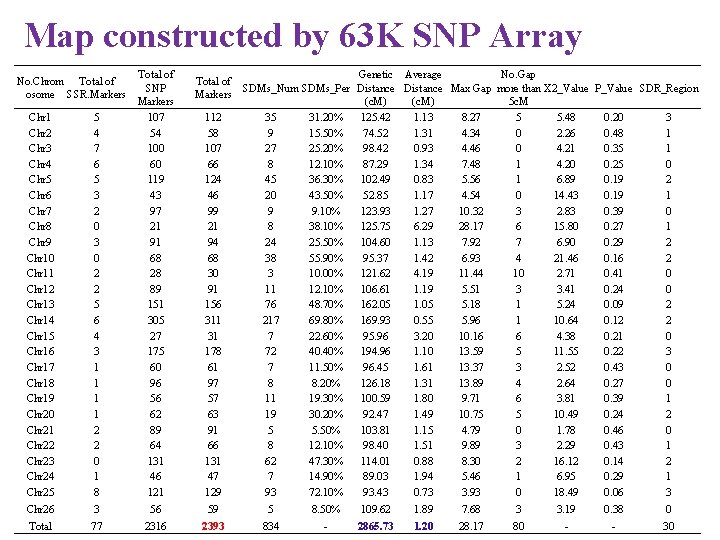
Map constructed by 63 K SNP Array Chr 1 Chr 2 Chr 3 Chr 4 Chr 5 Chr 6 Chr 7 Chr 8 Chr 9 Chr 10 Chr 11 Chr 12 Chr 13 Chr 14 Chr 15 Chr 16 Chr 17 Chr 18 Chr 19 Chr 20 Chr 21 Chr 22 Chr 23 Chr 24 Chr 25 5 4 7 6 5 3 2 0 3 0 2 2 5 6 4 3 1 1 2 2 0 1 8 Total of SNP Markers 107 54 100 60 119 43 97 21 91 68 28 89 151 305 27 175 60 96 56 62 89 64 131 46 121 Chr 26 3 56 59 5 8. 50% 109. 62 1. 89 7. 68 3 3. 19 0. 38 0 Total 77 2316 2393 834 - 2865. 73 1. 20 28. 17 80 - - 30 No. Chrom Total of osome SSR. Markers Total of Markers 112 58 107 66 124 46 99 21 94 68 30 91 156 311 31 178 61 97 57 63 91 66 131 47 129 Genetic Average No. Gap SDMs_Num SDMs_Per Distance Max Gap more than X 2_Value P_Value SDR_Region (c. M) 5 c. M 35 31. 20% 125. 42 1. 13 8. 27 5 5. 48 0. 20 3 9 15. 50% 74. 52 1. 31 4. 34 0 2. 26 0. 48 1 27 25. 20% 98. 42 0. 93 4. 46 0 4. 21 0. 35 1 8 12. 10% 87. 29 1. 34 7. 48 1 4. 20 0. 25 0 45 36. 30% 102. 49 0. 83 5. 56 1 6. 89 0. 19 2 20 43. 50% 52. 85 1. 17 4. 54 0 14. 43 0. 19 1 9 9. 10% 123. 93 1. 27 10. 32 3 2. 83 0. 39 0 8 38. 10% 125. 75 6. 29 28. 17 6 15. 80 0. 27 1 24 25. 50% 104. 60 1. 13 7. 92 7 6. 90 0. 29 2 38 55. 90% 95. 37 1. 42 6. 93 4 21. 46 0. 16 2 3 10. 00% 121. 62 4. 19 11. 44 10 2. 71 0. 41 0 11 12. 10% 106. 61 1. 19 5. 51 3 3. 41 0. 24 0 76 48. 70% 162. 05 1. 05 5. 18 1 5. 24 0. 09 2 217 69. 80% 169. 93 0. 55 5. 96 1 10. 64 0. 12 2 7 22. 60% 95. 96 3. 20 10. 16 6 4. 38 0. 21 0 72 40. 40% 194. 96 1. 10 13. 59 5 11. 55 0. 22 3 7 11. 50% 96. 45 1. 61 13. 37 3 2. 52 0. 43 0 8 8. 20% 126. 18 1. 31 13. 89 4 2. 64 0. 27 0 11 19. 30% 100. 59 1. 80 9. 71 6 3. 81 0. 39 1 19 30. 20% 92. 47 1. 49 10. 75 5 10. 49 0. 24 2 5 5. 50% 103. 81 1. 15 4. 79 0 1. 78 0. 46 0 8 12. 10% 98. 40 1. 51 9. 89 3 2. 29 0. 43 1 62 47. 30% 114. 01 0. 88 8. 30 2 16. 12 0. 14 2 7 14. 90% 89. 03 1. 94 5. 46 1 6. 95 0. 29 1 93 72. 10% 93. 43 0. 73 3. 93 0 18. 49 0. 06 3

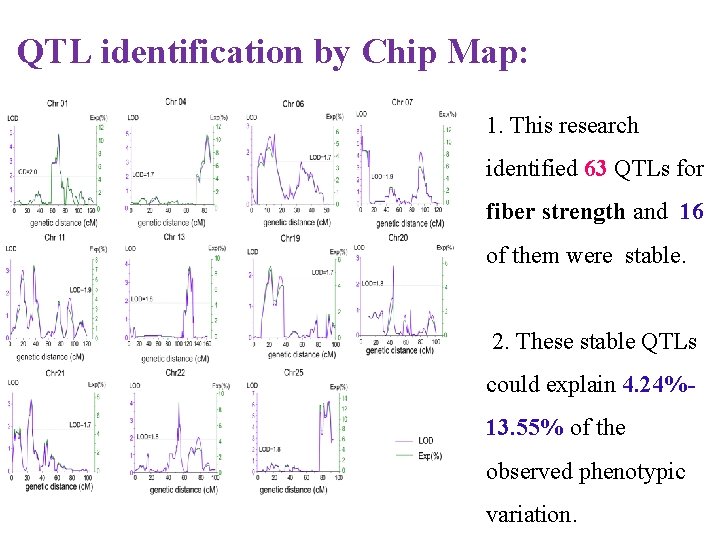
QTL identification by Chip Map: 1. This research identified 63 QTLs for fiber strength and 16 of them were stable. 2. These stable QTLs could explain 4. 24%13. 55% of the observed phenotypic variation.

Genetics map construction and QTL identification for boll weight with SNP markers discovered by SLAF-Seq Zhang zhen etal. BMC plant biology, 2016

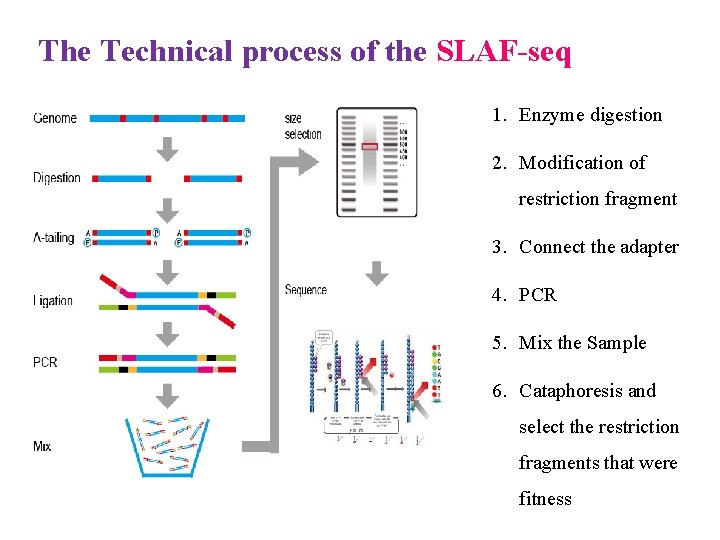
The Technical process of the SLAF-seq 1. Enzyme digestion 2. Modification of restriction fragment 3. Connect the adapter 4. PCR 5. Mix the Sample 6. Cataphoresis and select the restriction fragments that were fitness

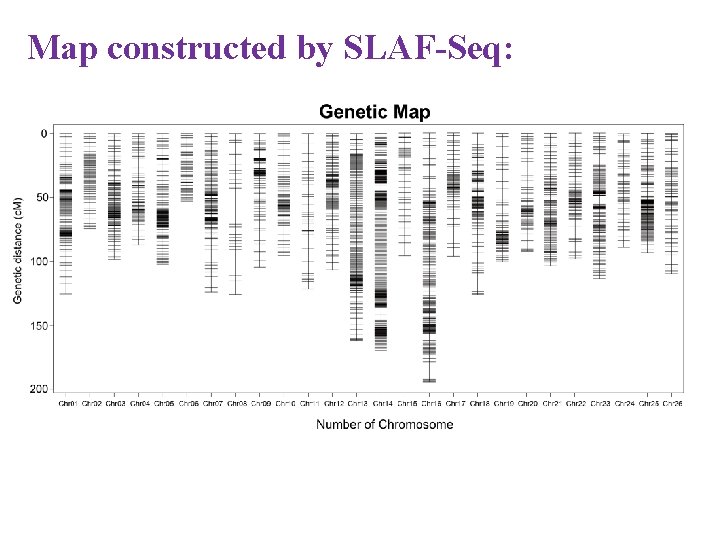
Map constructed by SLAF-Seq:

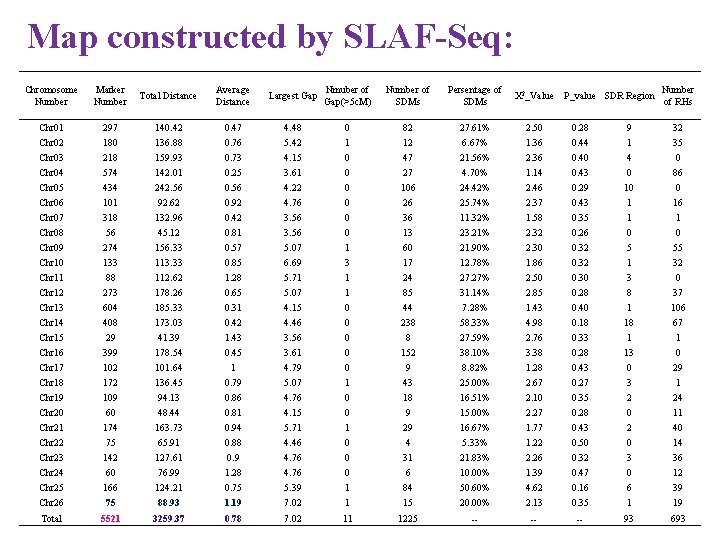
Map constructed by SLAF-Seq: Chromosome Number Marker Number Total Distance Average Distance Largest Gap Nmuber of Gap(>5 c. M) Number of SDMs Persentage of SDMs X 2_Value Chr 01 297 140. 42 0. 47 4. 48 0 82 27. 61% 2. 50 0. 28 9 32 Chr 02 180 136. 88 0. 76 5. 42 1 12 6. 67% 1. 36 0. 44 1 35 Chr 03 218 159. 93 0. 73 4. 15 0 47 21. 56% 2. 36 0. 40 4 0 Chr 04 574 142. 01 0. 25 3. 61 0 27 4. 70% 1. 14 0. 43 0 86 Chr 05 434 242. 56 0. 56 4. 22 0 106 24. 42% 2. 46 0. 29 10 0 Chr 06 101 92. 62 0. 92 4. 76 0 26 25. 74% 2. 37 0. 43 1 16 Chr 07 318 132. 96 0. 42 3. 56 0 36 11. 32% 1. 58 0. 35 1 1 Chr 08 56 45. 12 0. 81 3. 56 0 13 23. 21% 2. 32 0. 26 0 0 Chr 09 274 156. 33 0. 57 5. 07 1 60 21. 90% 2. 30 0. 32 5 55 Chr 10 133 113. 33 0. 85 6. 69 3 17 12. 78% 1. 86 0. 32 1 32 Chr 11 88 112. 62 1. 28 5. 71 1 24 27. 27% 2. 50 0. 30 3 0 Chr 12 273 178. 26 0. 65 5. 07 1 85 31. 14% 2. 85 0. 28 8 37 Chr 13 604 185. 33 0. 31 4. 15 0 44 7. 28% 1. 43 0. 40 1 106 Chr 14 408 173. 03 0. 42 4. 46 0 238 58. 33% 4. 98 0. 18 18 67 Chr 15 29 41. 39 1. 43 3. 56 0 8 27. 59% 2. 76 0. 33 1 1 Chr 16 399 178. 54 0. 45 3. 61 0 152 38. 10% 3. 38 0. 28 13 0 Chr 17 102 101. 64 1 4. 79 0 9 8. 82% 1. 28 0. 43 0 29 Chr 18 172 136. 45 0. 79 5. 07 1 43 25. 00% 2. 67 0. 27 3 1 Chr 19 109 94. 13 0. 86 4. 76 0 18 16. 51% 2. 10 0. 35 2 24 P_value SDR Region Number of RHs Chr 20 60 48. 44 0. 81 4. 15 0 9 15. 00% 2. 27 0. 28 0 11 Chr 21 174 163. 73 0. 94 5. 71 1 29 16. 67% 1. 77 0. 43 2 40 Chr 22 75 65. 91 0. 88 4. 46 0 4 5. 33% 1. 22 0. 50 0 14 Chr 23 142 127. 61 0. 9 4. 76 0 31 21. 83% 2. 26 0. 32 3 36 Chr 24 60 76. 99 1. 28 4. 76 0 6 10. 00% 1. 39 0. 47 0 12 Chr 25 166 124. 21 0. 75 5. 39 1 84 50. 60% 4. 62 0. 16 6 39 Chr 26 75 88. 93 1. 19 7. 02 1 15 20. 00% 2. 13 0. 35 1 19 Total 5521 3259. 37 0. 78 7. 02 11 1225 -- -- -- 93 693

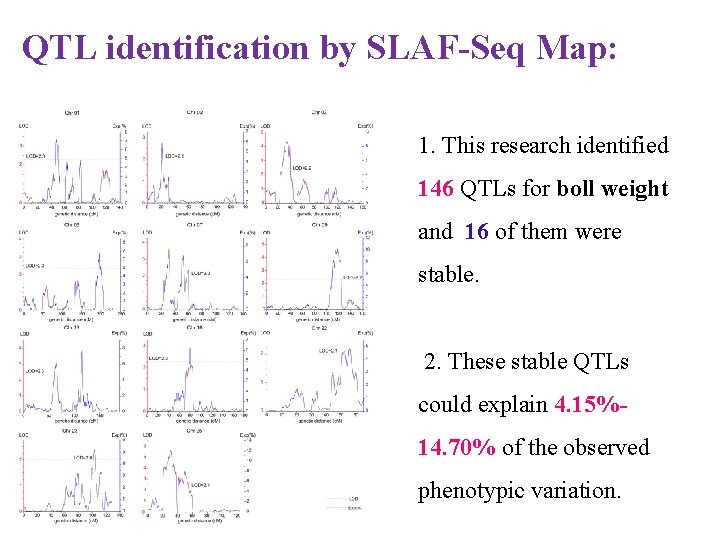
QTL identification by SLAF-Seq Map: 1. This research identified 146 QTLs for boll weight and 16 of them were stable. 2. These stable QTLs could explain 4. 15%14. 70% of the observed phenotypic variation.

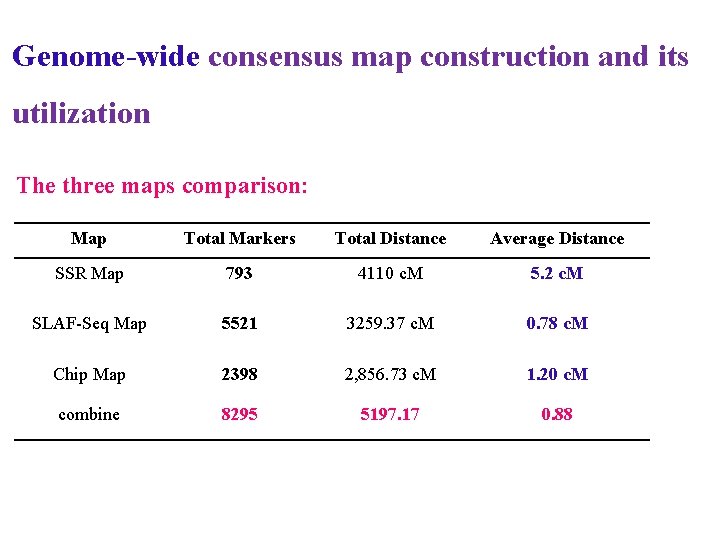
Genome-wide consensus map construction and its utilization The three maps comparison: Map Total Markers Total Distance Average Distance SSR Map 793 4110 c. M 5. 2 c. M SLAF-Seq Map 5521 3259. 37 c. M 0. 78 c. M Chip Map 2398 2, 856. 73 c. M 1. 20 c. M combine 8295 5197. 17 0. 88

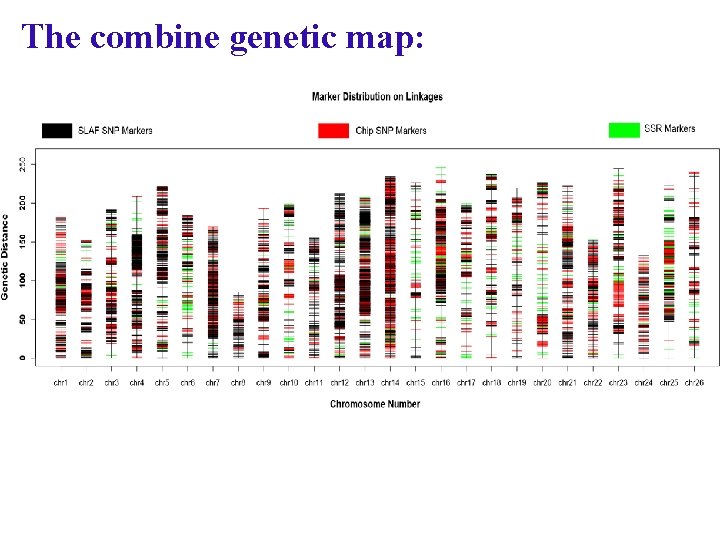
The combine genetic map:

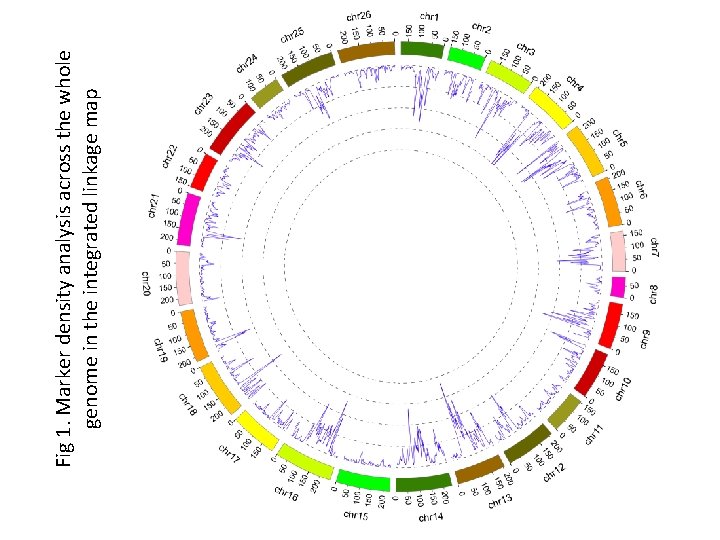
Fig 1. Marker density analysis across the whole genome in the integrated linkage map

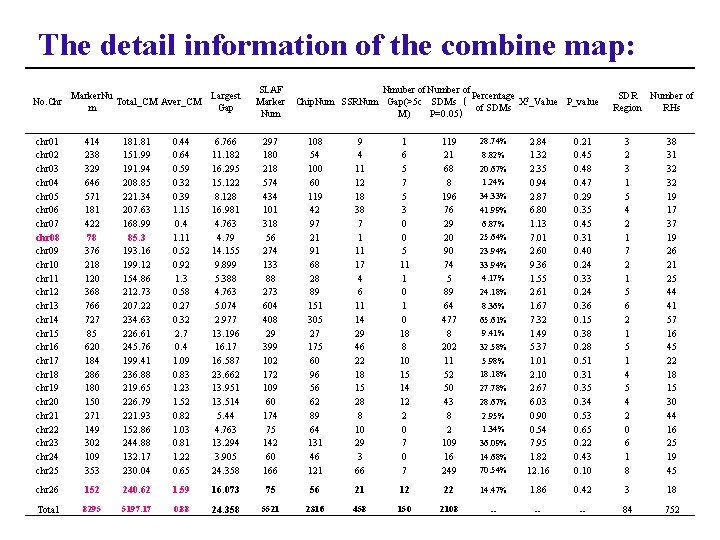
The detail information of the combine map: No. Chr Marker. Nu Largest Total_CM Aver_CM m Gap SLAF Marker Num Nmuber of Number of Percentage 2 Chip. Num SSRNum Gap(>5 c SDMs( X _Value P_value of SDMs M) P=0. 05) chr 01 chr 02 chr 03 chr 04 chr 05 chr 06 chr 07 chr 08 chr 09 chr 10 chr 11 chr 12 chr 13 chr 14 chr 15 chr 16 chr 17 chr 18 chr 19 chr 20 chr 21 chr 22 chr 23 chr 24 chr 25 414 238 329 646 571 181 422 78 376 218 120 368 766 727 85 620 184 286 180 150 271 149 302 109 353 181. 81 151. 99 191. 94 208. 85 221. 34 207. 63 168. 99 85. 3 193. 16 199. 12 154. 86 212. 73 207. 22 234. 63 226. 61 245. 76 199. 41 236. 88 219. 65 226. 79 221. 93 152. 86 244. 88 132. 17 230. 04 0. 44 0. 64 0. 59 0. 32 0. 39 1. 15 0. 4 1. 11 0. 52 0. 92 1. 3 0. 58 0. 27 0. 32 2. 7 0. 4 1. 09 0. 83 1. 23 1. 52 0. 82 1. 03 0. 81 1. 22 0. 65 6. 766 11. 182 16. 295 15. 122 8. 128 16. 981 4. 763 4. 79 14. 155 9. 899 5. 388 4. 763 5. 074 2. 977 13. 196 16. 17 16. 587 23. 662 13. 951 13. 514 5. 44 4. 763 13. 294 3. 905 24. 358 297 180 218 574 434 101 318 56 274 133 88 273 604 408 29 399 102 172 109 60 174 75 142 60 166 108 54 100 60 119 42 97 21 91 68 28 89 151 305 27 175 60 96 56 62 89 64 131 46 121 9 4 11 12 18 38 7 1 11 17 4 6 11 14 29 46 22 18 15 28 8 10 29 3 66 1 6 5 7 5 3 0 0 5 11 1 0 18 8 10 15 14 12 2 0 7 119 21 68 8 196 76 29 20 90 74 5 89 64 477 8 202 11 52 50 43 8 2 109 16 249 28. 74% chr 26 152 240. 62 1. 59 16. 073 75 56 21 12 Total 8295 5197. 17 0. 88 24. 358 5521 2316 458 150 SDR Number of Region RHs 70. 54% 2. 84 1. 32 2. 35 0. 94 2. 87 6. 80 1. 13 7. 01 2. 60 9. 36 1. 55 2. 61 1. 67 7. 32 1. 49 5. 37 1. 01 2. 10 2. 67 6. 03 0. 90 0. 54 7. 95 1. 82 12. 16 0. 21 0. 45 0. 48 0. 47 0. 29 0. 35 0. 45 0. 31 0. 40 0. 24 0. 33 0. 24 0. 36 0. 15 0. 38 0. 28 0. 51 0. 35 0. 34 0. 53 0. 65 0. 22 0. 43 0. 10 3 2 3 1 5 4 2 1 7 2 1 5 6 2 1 5 1 4 5 4 2 0 6 1 8 38 31 32 32 19 17 37 19 26 21 25 44 41 57 16 45 22 18 15 30 44 16 25 19 45 22 14. 47% 1. 86 0. 42 3 18 2108 -- -- -- 84 752 8. 82% 20. 67% 1. 24% 34. 33% 41. 99% 6. 87% 25. 64% 23. 94% 33. 94% 4. 17% 24. 18% 8. 36% 65. 61% 9. 41% 32. 58% 5. 98% 18. 18% 27. 78% 28. 67% 2. 95% 1. 34% 36. 09% 14. 68%

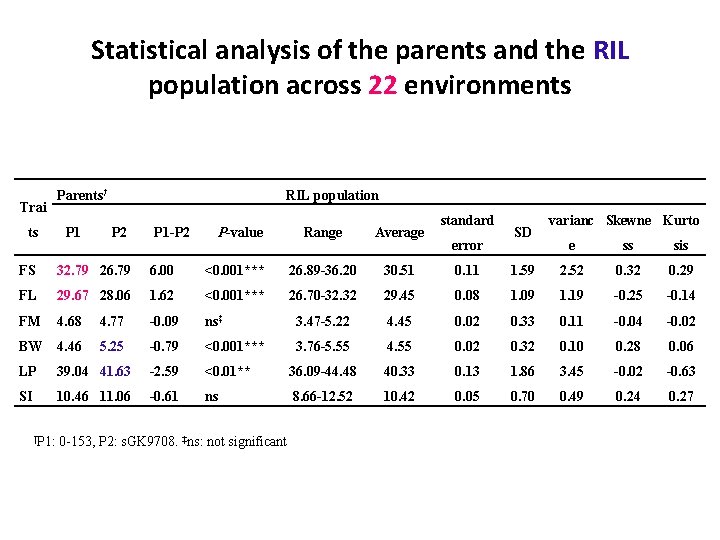
Statistical analysis of the parents and the RIL population across 22 environments Trai ts Parents† P 1 RIL population P 2 P 1 -P 2 P-value Range Average standard error SD varianc Skewne Kurto e ss sis FS 32. 79 26. 79 6. 00 <0. 001*** 26. 89 -36. 20 30. 51 0. 11 1. 59 2. 52 0. 32 0. 29 FL 29. 67 28. 06 1. 62 <0. 001*** 26. 70 -32. 32 29. 45 0. 08 1. 09 1. 19 -0. 25 -0. 14 FM 4. 68 4. 77 -0. 09 ns‡ 3. 47 -5. 22 4. 45 0. 02 0. 33 0. 11 -0. 04 -0. 02 BW 4. 46 5. 25 -0. 79 <0. 001*** 3. 76 -5. 55 4. 55 0. 02 0. 32 0. 10 0. 28 0. 06 LP 39. 04 41. 63 -2. 59 <0. 01** 36. 09 -44. 48 40. 33 0. 13 1. 86 3. 45 -0. 02 -0. 63 SI 10. 46 11. 06 -0. 61 ns 8. 66 -12. 52 10. 42 0. 05 0. 70 0. 49 0. 24 0. 27 †P 1: 0 -153, P 2: s. GK 9708. ‡ns: not significant

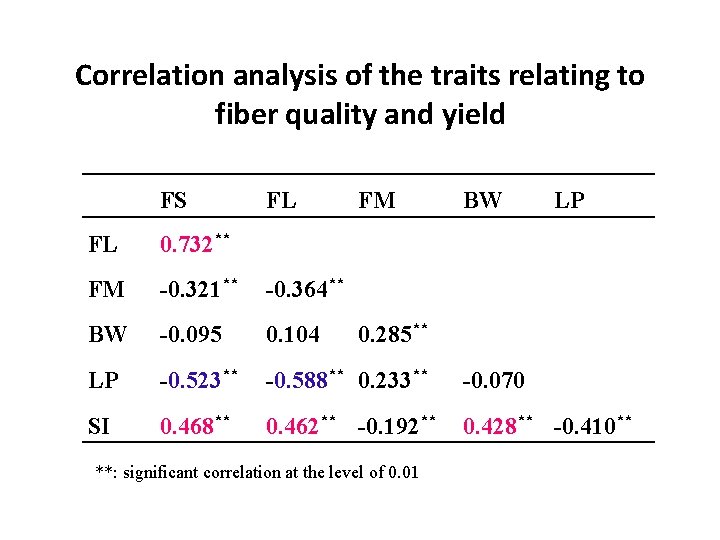
Correlation analysis of the traits relating to fiber quality and yield FS FL FM BW LP FL 0. 732** FM -0. 321** -0. 364** BW -0. 095 0. 104 LP -0. 523** -0. 588** 0. 233** -0. 070 SI 0. 468** 0. 462** -0. 192** 0. 428** -0. 410** 0. 285** **: significant correlation at the level of 0. 01

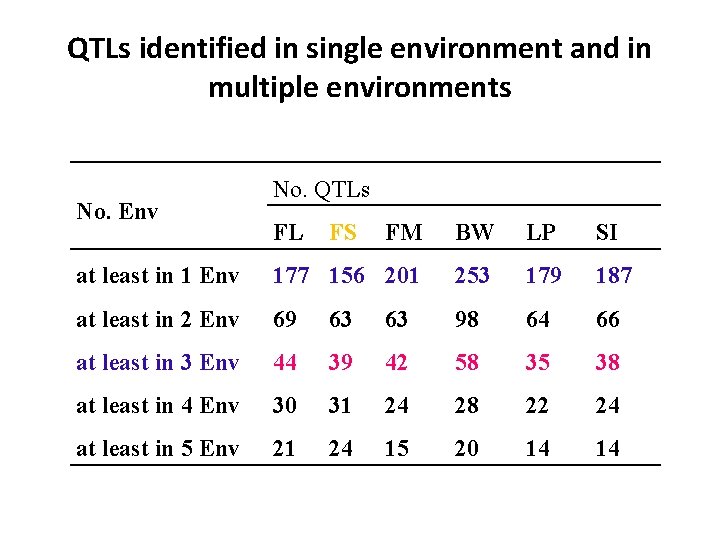
QTLs identified in single environment and in multiple environments No. Env No. QTLs FL FS FM BW LP SI at least in 1 Env 177 156 201 253 179 187 at least in 2 Env 69 63 63 98 64 66 at least in 3 Env 44 39 42 58 35 38 at least in 4 Env 30 31 24 28 22 24 at least in 5 Env 21 24 15 20 14 14

QTL identification and distribution

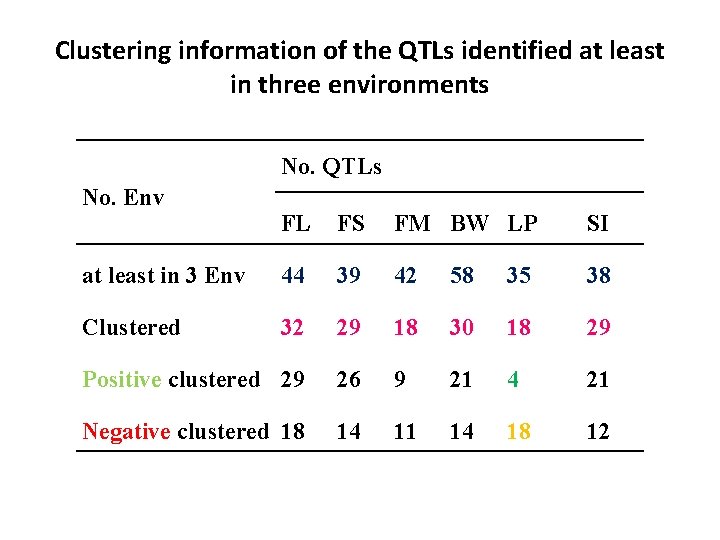
Clustering information of the QTLs identified at least in three environments No. QTLs No. Env FL FS FM BW LP SI at least in 3 Env 44 39 42 58 35 38 Clustered 32 29 18 30 18 29 Positive clustered 29 26 9 21 4 21 Negative clustered 18 14 11 14 18 12

Summery of QTL Clusters

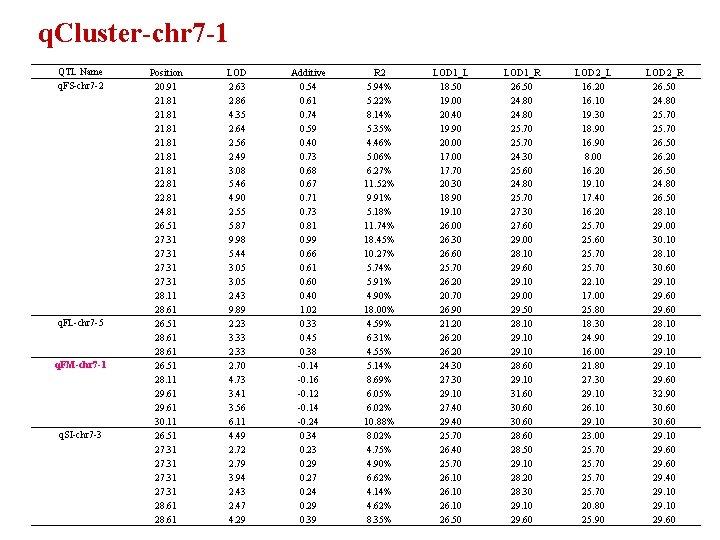
q. Cluster-chr 7 -1 QTL Name q. FS-chr 7 -2 q. FL-chr 7 -5 q. FM-chr 7 -1 q. SI-chr 7 -3 Position 20. 91 21. 81 22. 81 24. 81 26. 51 27. 31 28. 11 28. 61 26. 51 28. 11 29. 61 30. 11 26. 51 27. 31 28. 61 LOD 2. 63 2. 86 4. 35 2. 64 2. 56 2. 49 3. 08 5. 46 4. 90 2. 55 5. 87 9. 98 5. 44 3. 05 2. 43 9. 89 2. 23 3. 33 2. 70 4. 73 3. 41 3. 56 6. 11 4. 49 2. 72 2. 79 3. 94 2. 43 2. 47 4. 29 Additive 0. 54 0. 61 0. 74 0. 59 0. 40 0. 73 0. 68 0. 67 0. 71 0. 73 0. 81 0. 99 0. 66 0. 61 0. 60 0. 40 1. 02 0. 33 0. 45 0. 38 -0. 14 -0. 16 -0. 12 -0. 14 -0. 24 0. 34 0. 23 0. 29 0. 27 0. 24 0. 29 0. 39 R 2 5. 94% 5. 22% 8. 14% 5. 35% 4. 46% 5. 06% 6. 27% 11. 52% 9. 91% 5. 18% 11. 74% 18. 45% 10. 27% 5. 74% 5. 91% 4. 90% 18. 00% 4. 59% 6. 31% 4. 55% 5. 14% 8. 69% 6. 05% 6. 02% 10. 88% 8. 02% 4. 75% 4. 90% 6. 62% 4. 14% 4. 62% 8. 35% LOD 1_L 18. 50 19. 00 20. 40 19. 90 20. 00 17. 70 20. 30 18. 90 19. 10 26. 00 26. 30 26. 60 25. 70 26. 20 20. 70 26. 90 21. 20 26. 20 24. 30 27. 30 29. 10 27. 40 29. 40 25. 70 26. 10 26. 50 LOD 1_R 26. 50 24. 80 25. 70 24. 30 25. 60 24. 80 25. 70 27. 30 27. 60 29. 00 28. 10 29. 60 29. 10 29. 00 29. 50 28. 10 29. 10 28. 60 29. 10 31. 60 30. 60 28. 50 29. 10 28. 20 28. 30 29. 10 29. 60 LOD 2_L 16. 20 16. 10 19. 30 18. 90 16. 90 8. 00 16. 20 19. 10 17. 40 16. 20 25. 70 25. 60 25. 70 22. 10 17. 00 25. 80 18. 30 24. 90 16. 00 21. 80 27. 30 29. 10 26. 10 29. 10 23. 00 25. 70 20. 80 25. 90 LOD 2_R 26. 50 24. 80 25. 70 26. 50 26. 20 26. 50 24. 80 26. 50 28. 10 29. 00 30. 10 28. 10 30. 60 29. 10 29. 60 28. 10 29. 60 32. 90 30. 60 29. 10 29. 60 29. 40 29. 10 29. 60

q. Cluster-chr 25 -1 QTL Name Position LOD Additive R 2 LOD 1_L LOD 1_R LOD 2_L LOD 2_R q. FS-chr 25 -2 111. 41 2. 48 0. 57 4. 78% 108. 60 112. 20 106. 60 112. 20 114. 41 2. 62 0. 63 4. 88% 112. 90 119. 60 112. 90 121. 70 114. 41 4. 53 0. 76 9. 32% 113. 90 115. 80 112. 70 118. 90 116. 31 3. 45 0. 76 6. 06% 115. 40 118. 80 115. 40 120. 00 q. FL-chr 25 -7 115. 41 3. 92 0. 42 9. 14% 114. 20 118. 40 114. 50 119. 60 116. 31 3. 27 0. 51 7. 13% 115. 40 119. 50 112. 90 120. 00 116. 31 6. 37 0. 83 13. 48% 115. 40 118. 70 115. 40 119. 50 116. 31 3. 41 0. 55 7. 51% 115. 40 119. 30 112. 90 120. 90 116. 31 2. 97 0. 43 6. 16% 115. 20 120. 90 112. 90 121. 70 116. 31 3. 05 0. 33 6. 49% 114. 30 119. 70 112. 90 121. 70 117. 31 5. 26 0. 66 12. 33% 115. 90 119. 30 112. 90 120. 00 117. 31 3. 08 0. 48 6. 78% 116. 30 119. 60 116. 30 121. 20 q. LP-chr 25 -7 115. 41 3. 86 -0. 66 8. 22% 112. 90 118. 60 112. 90 120. 60 115. 41 3. 37 -0. 66 7. 72% 112. 90 119. 30 112. 90 120. 20 116. 31 4. 14 -0. 84 8. 56% 113. 50 119. 60 112. 40 120. 90 116. 31 4. 52 -0. 73 9. 21% 114. 10 120. 00 112. 90 121. 00 117. 31 4. 01 -1. 27 9. 26% 116. 20 118. 30 115. 80 119. 40 117. 31 4. 08 -0. 61 9. 05% 116. 50 119. 30 115. 40 120. 20 117. 31 3. 37 -0. 62 7. 20% 116. 30 120. 00 116. 30 121. 80 119. 31 2. 60 -0. 67 5. 81% 115. 00 122. 20 112. 90 122. 20 119. 31 3. 18 -0. 68 7. 22% 117. 30 121. 60 117. 30 122. 20 q. BW-chr 25 -11 119. 31 3. 89 -0. 13 8. 52% 117. 60 121. 40 116. 90 122. 60 119. 31 3. 90 -0. 15 8. 32% 118. 20 122. 70 116. 30 123. 70 119. 31 6. 92 -0. 20 14. 66% 118. 00 121. 50 119. 20 122. 20 119. 31 6. 08 -0. 20 14. 08% 118. 20 121. 80 117. 50 122. 80

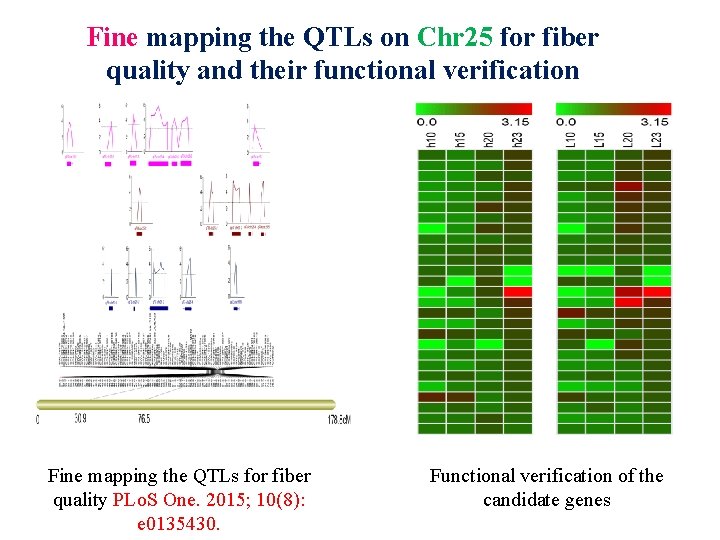
Fine mapping the QTLs on Chr 25 for fiber quality and their functional verification Fine mapping the QTLs for fiber quality PLo. S One. 2015; 10(8): e 0135430. Functional verification of the candidate genes

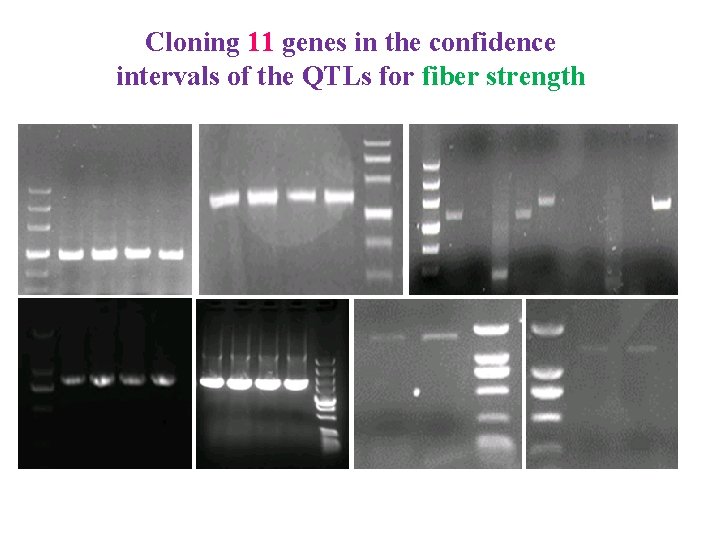
Cloning 11 genes in the confidence intervals of the QTLs for fiber strength

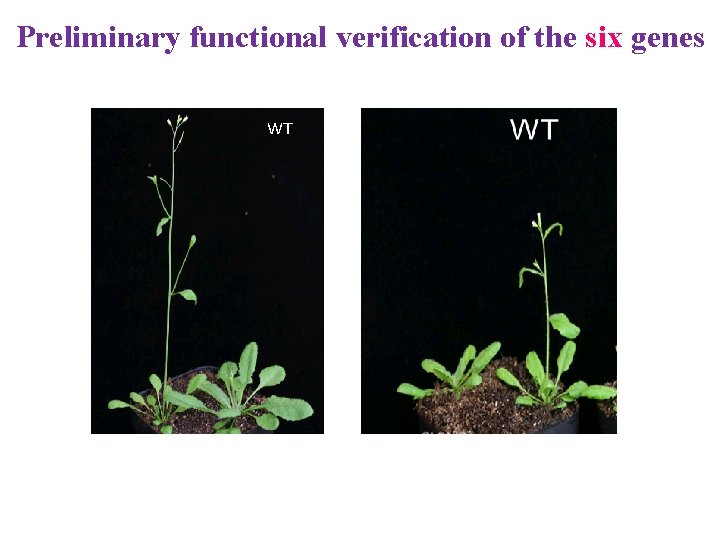
Preliminary functional verification of the six genes WT

THANK YOU!