CONCERTS Dynamic Connection of Fragments as an Approach

- Slides: 17

CONCERTS: Dynamic Connection of Fragments as an Approach to de Novo Ligand Design Creation Of Novel Compounds by Evaluation of Residues at Target Sites David A. Pearlman and Mark A. Murkco Vertex Pharmaceuticals Incorperated Cambridge, MA 1

Outline ● Background ● Implementation ● HIV-1 aspartyl protease ● FK 506 binding protein ● Conclusions 2

Previous Work: CONCEPTS ● Active site is filled with atoms ● Run MD simulations, and form/break bonds ● Generates useful de Novo leads ● Limitations Difficult to incorporate charge models – Slow convergence, especially for “spacer” regions – Only 1 suggestion per cpu-intensive run – 3

CONCERTS: Implementation Modified AMBER/SANDER 4. 0 minimization/MD program 1) Active site is filled with user-defined fragments 2) “Connection vectors” are chosen for each fragment 3) Define a volume for a known protein of interest 4) Randomly orient fragments in defined volume 5) Fragment minimization and MD (two steps) 6) Start CONCERTS 4

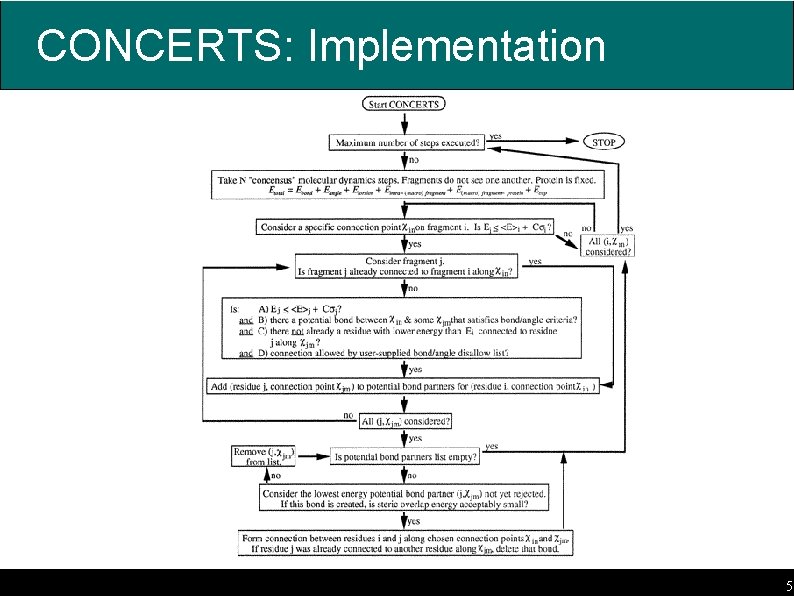
CONCERTS: Implementation 5

CONCERTS: Improvements CONCERTS has several improvements over CONCEPTS: ● ● Fragments can inherently have charge Fragments span larger region of space; don't have to worry about “spacer” regions Many suggested molecules can be built during a run Greater control over types of molecules generated 6

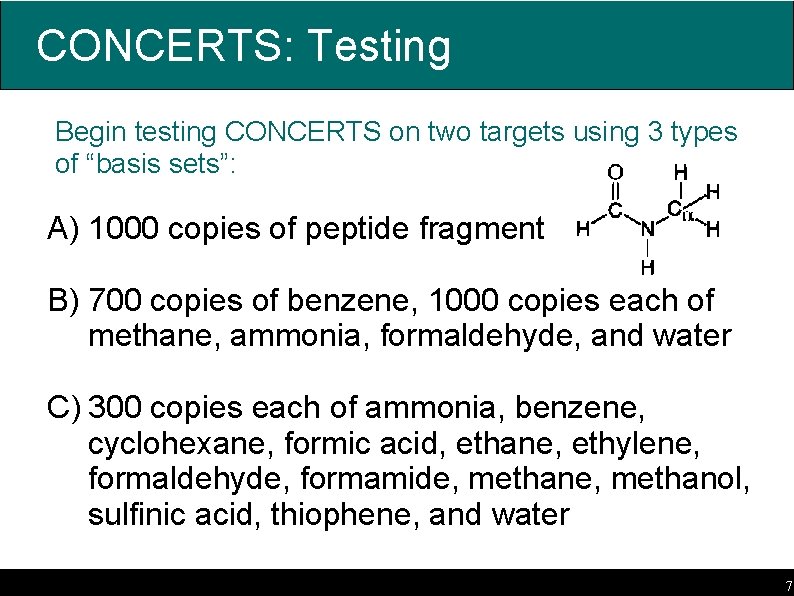
CONCERTS: Testing Begin testing CONCERTS on two targets using 3 types of “basis sets”: A) 1000 copies of peptide fragment B) 700 copies of benzene, 1000 copies each of methane, ammonia, formaldehyde, and water C) 300 copies each of ammonia, benzene, cyclohexane, formic acid, ethane, ethylene, formaldehyde, formamide, methanol, sulfinic acid, thiophene, and water 7

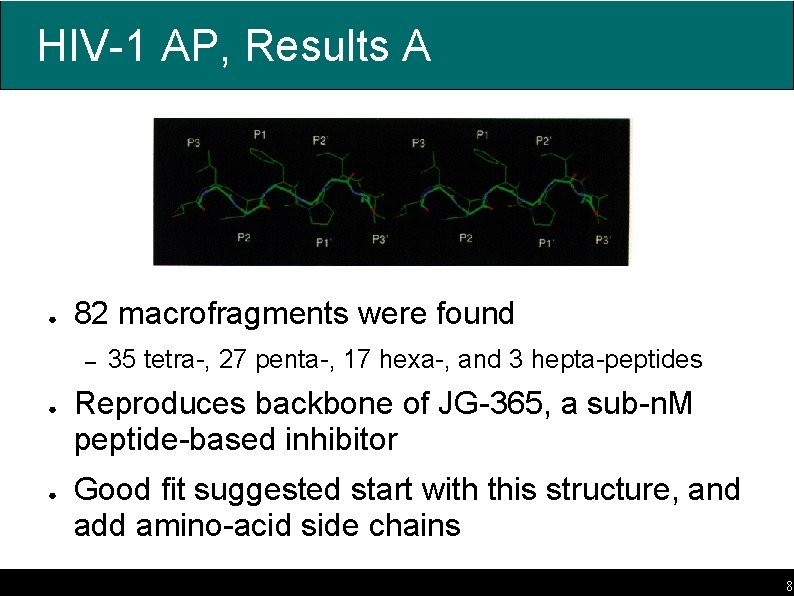
HIV-1 AP, Results A ● 82 macrofragments were found – ● ● 35 tetra-, 27 penta-, 17 hexa-, and 3 hepta-peptides Reproduces backbone of JG-365, a sub-n. M peptide-based inhibitor Good fit suggested start with this structure, and add amino-acid side chains 8

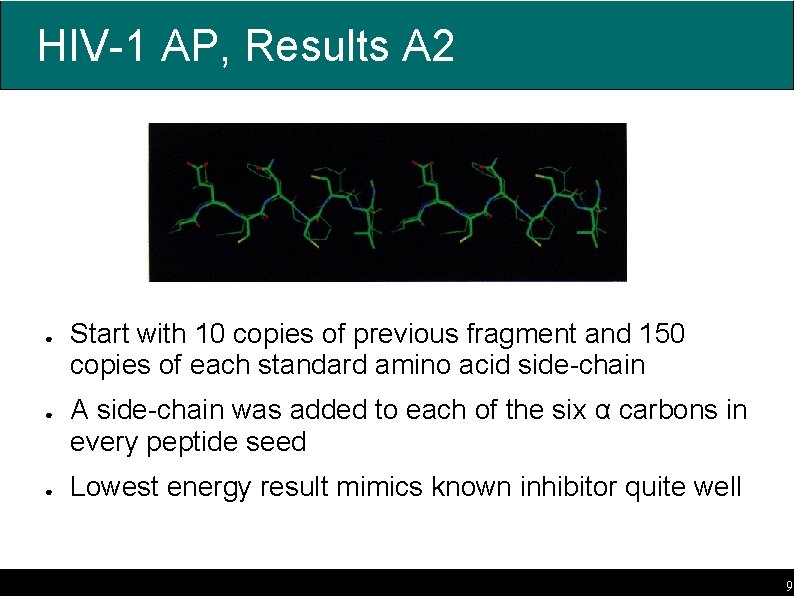
HIV-1 AP, Results A 2 ● ● ● Start with 10 copies of previous fragment and 150 copies of each standard amino acid side-chain A side-chain was added to each of the six α carbons in every peptide seed Lowest energy result mimics known inhibitor quite well 9

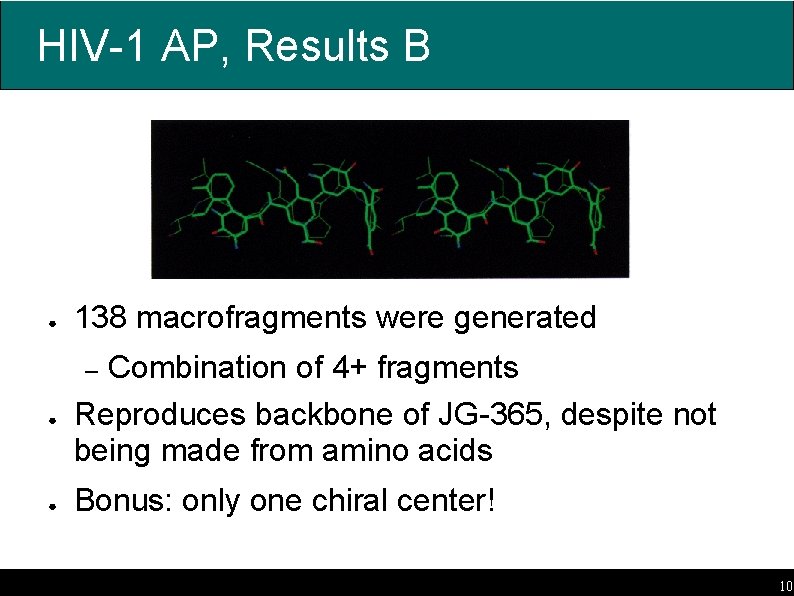
HIV-1 AP, Results B ● 138 macrofragments were generated Combination of 4+ fragments Reproduces backbone of JG-365, despite not being made from amino acids – ● ● Bonus: only one chiral center! 10

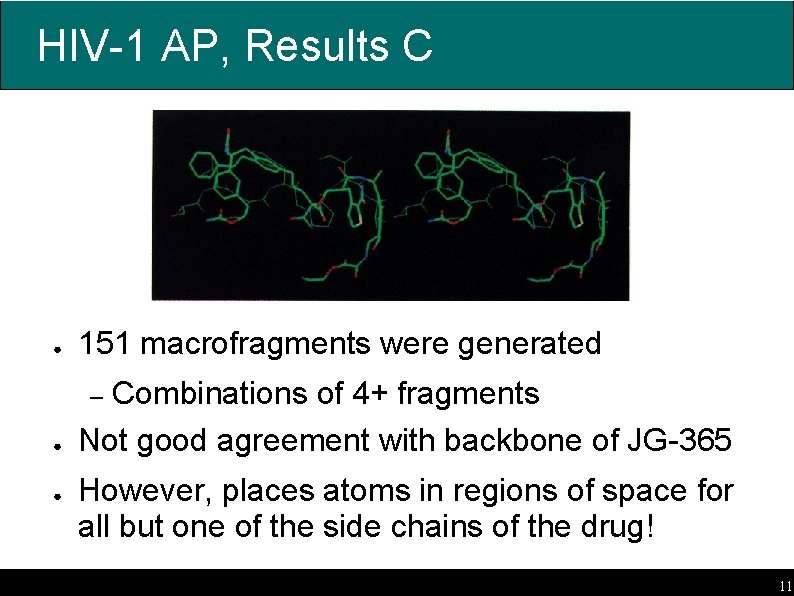
HIV-1 AP, Results C ● 151 macrofragments were generated ● Combinations of 4+ fragments Not good agreement with backbone of JG-365 – ● However, places atoms in regions of space for all but one of the side chains of the drug! 11

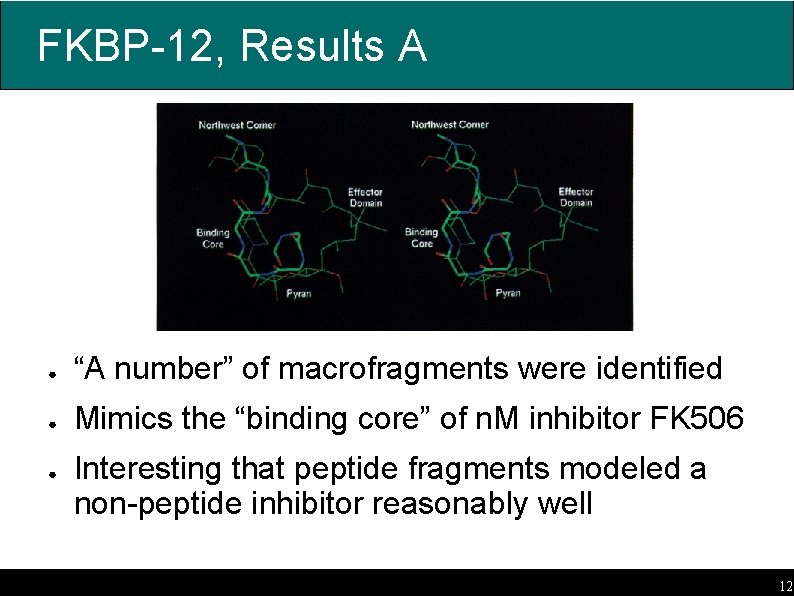
FKBP-12, Results A ● “A number” of macrofragments were identified ● Mimics the “binding core” of n. M inhibitor FK 506 ● Interesting that peptide fragments modeled a non-peptide inhibitor reasonably well 12

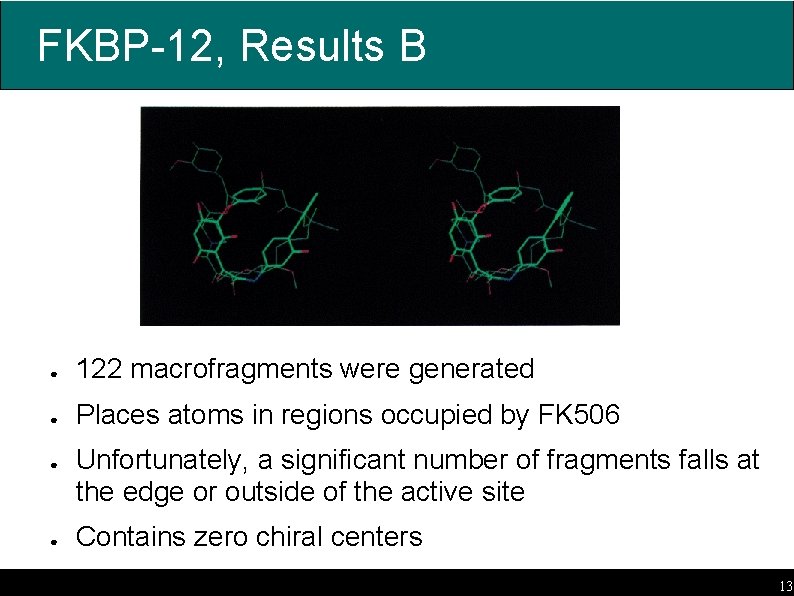
FKBP-12, Results B ● 122 macrofragments were generated ● Places atoms in regions occupied by FK 506 ● ● Unfortunately, a significant number of fragments falls at the edge or outside of the active site Contains zero chiral centers 13

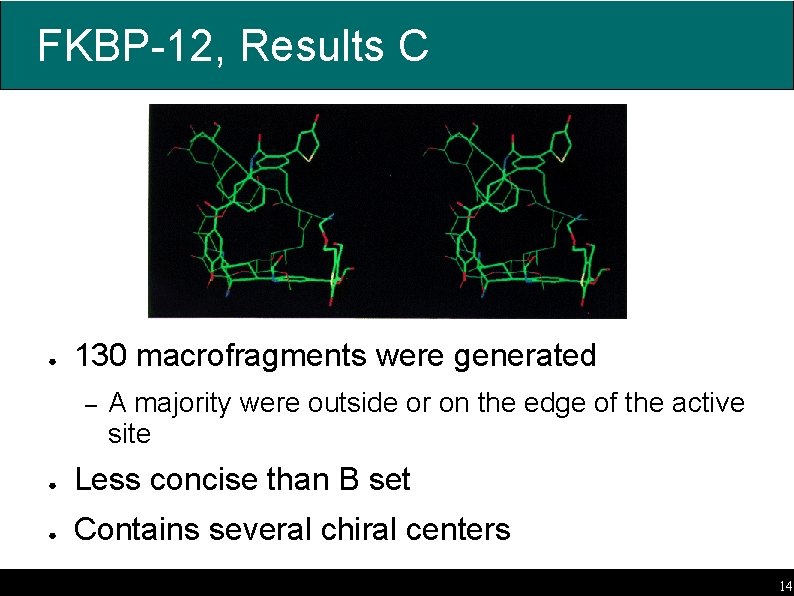
FKBP-12, Results C ● 130 macrofragments were generated – A majority were outside or on the edge of the active site ● Less concise than B set ● Contains several chiral centers 14

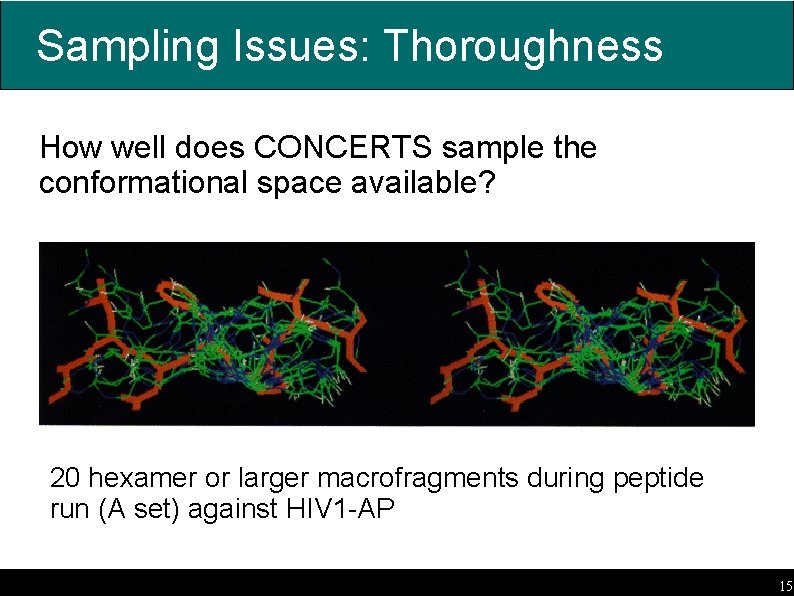
Sampling Issues: Thoroughness How well does CONCERTS sample the conformational space available? 20 hexamer or larger macrofragments during peptide run (A set) against HIV 1 -AP 15

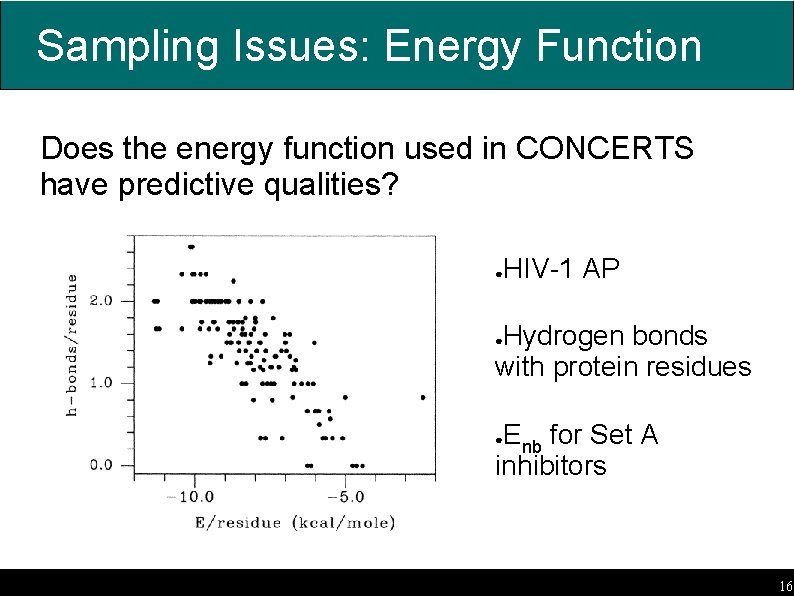
Sampling Issues: Energy Function Does the energy function used in CONCERTS have predictive qualities? HIV-1 AP ● Hydrogen bonds with protein residues ● Enb for Set A inhibitors ● 16

Conclusion ● CONCERTS works: it generates inhibitors – ● ● Peptide fragments produce more structures that are similar to known inhibitors More fragment types lead to increased diversity, but often have less similarity to inhibitors – ● ● For two targets: HIV-1 protease and FKBP-12 However, could produce new lead structures Less diverse fragment sets results in greater “convergence” For targets with unknown inhibitors, multiple structures can be generated – Identify trends or new leads for better modeling 17