Computer simulations in drug design and the GRID




- Slides: 34

Computer simulations in drug design, and the GRID Dr Jonathan W Essex University of Southampton

Computer simulations? • Molecular dynamics – Solve F = ma for particles in system – Follow time evolution of system – Average system properties over time • Monte Carlo – Perform random moves on system – Accept or reject move on the basis of change in energy – No time information

Drug action • Administration Free energy simulations – Oral, intravenous… – p. Ka, solubility • Transport and permeation – Passage through membranes – Partition coefficient, permeability • Protein binding – Binding affinity, specificity – Allosteric effects • Metabolism – P 450 enzymes – Catalysis Digital filtering Parallel tempering Free energy simulations Free QM/MM energy simulations Free energy simulations Structure generation Modelling

Three main areas of work • Protein-ligand binding – Structure prediction – Binding affinity prediction • Membrane modelling and small molecule permeation – Bioavailability – Coarse-grained membrane models • Protein conformational change – Induced-fit – Large-scale conformational change

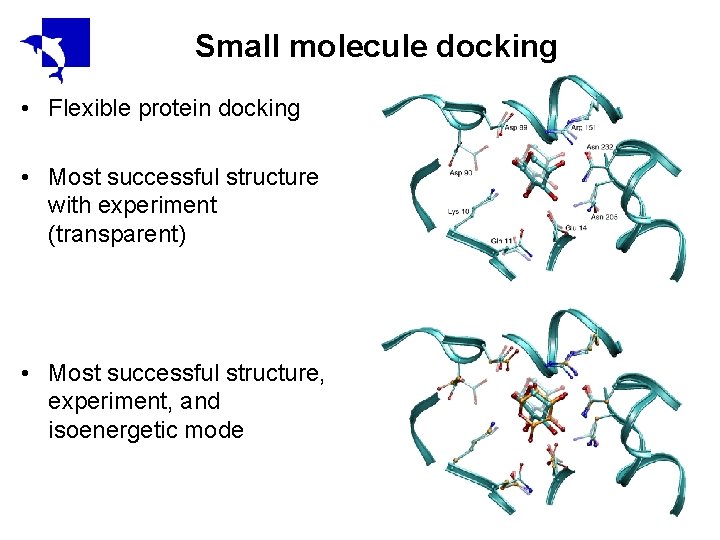
Small molecule docking • Flexible protein docking • Most successful structure with experiment (transparent) • Most successful structure, experiment, and isoenergetic mode

Replica-exchange free energy • Free energy methods that are applied between exchanges are the same as normal • Exchanges require little extra computational cost

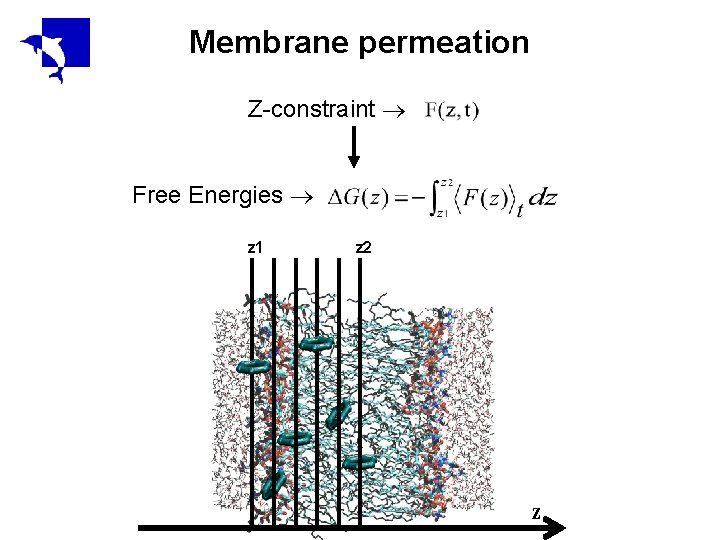
Membrane permeation Z-constraint Free Energies z 1 z 2 z

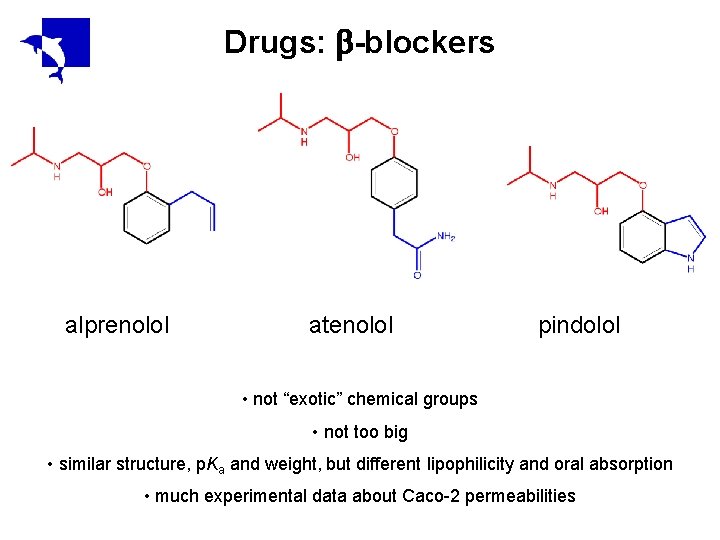
Drugs: -blockers alprenolol atenolol pindolol • not “exotic” chemical groups • not too big • similar structure, p. Ka and weight, but different lipophilicity and oral absorption • much experimental data about Caco-2 permeabilities

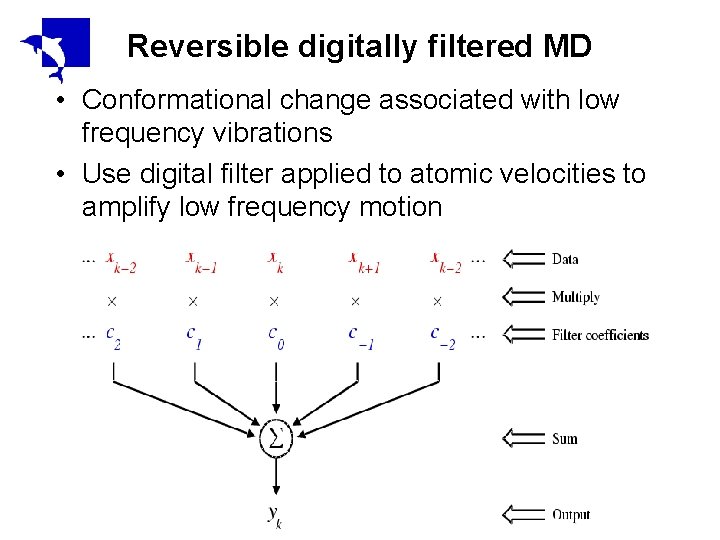
Reversible digitally filtered MD • Conformational change associated with low frequency vibrations • Use digital filter applied to atomic velocities to amplify low frequency motion

RDFMD • Effect of a quenching filter

Comb-e-Chem • EPSRC Grid computing project • Main objectives: – Equipment on the Grid – high-throughput small molecule crystallography – E-lab – pervasive computing, provenance – Structure modelling and property prediction • Automatic calculations performed on deposition of a new structure to the database • Distributed (Grid) computing

Distributed computing • • Heterogeneous computers – personal PCs Rather unstable Network often slow Suited for embarrassingly parallel calculations – SETI@Home, Folding@Home etc. – E-malaria (JISC) • Poor-man’s resource – Cheap – Large amount of power available

Condor • Condor project started in 1988 to harness the spare cycles of desktop computers • Available as a free download from the University of Wisconsin-Madison • Runs on Linux, Unix and Windows. Unix software can be compiled for Windows using Cygwin • For more information see http: //www. cs. wisc. edu/condor/

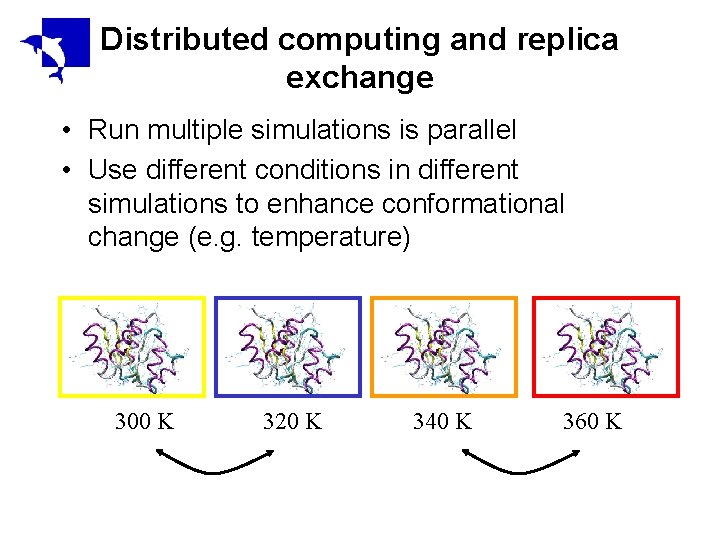
Distributed computing and replica exchange • Run multiple simulations is parallel • Use different conditions in different simulations to enhance conformational change (e. g. temperature) 300 K 320 K 340 K 360 K

300 K 320 K 340 K 360 K 380 K 400 K

300 K 1 320 K 340 K 360 K 380 K 400 K

300 K 320 K 340 K 360 K 380 K 400 K 1 1 1

300 K 320 K 340 K 360 K 380 K 400 K 2 2 2

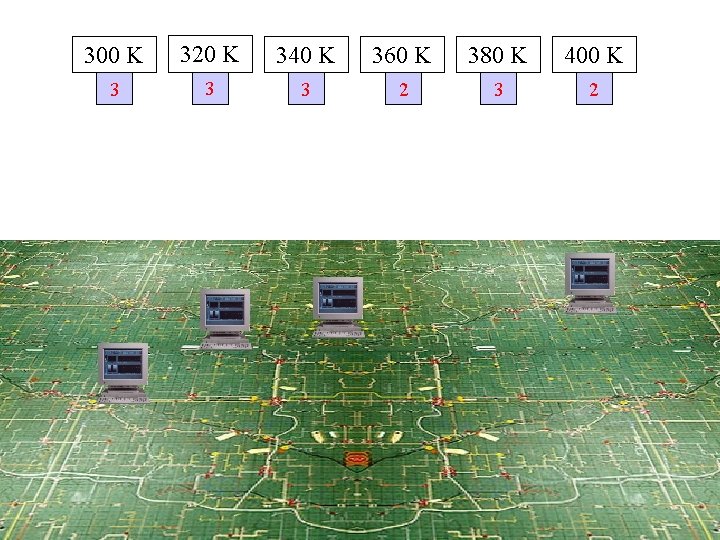
300 K 320 K 340 K 360 K 380 K 400 K 3 3 3 2

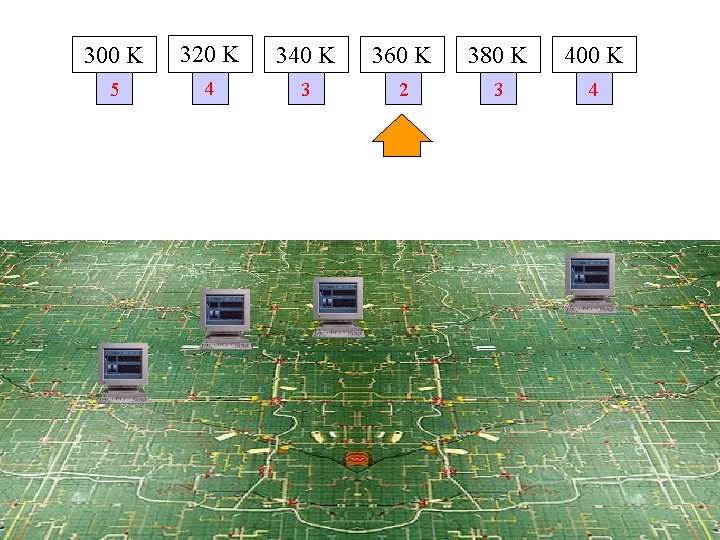
300 K 320 K 340 K 360 K 380 K 400 K 5 4 3 2 3 4

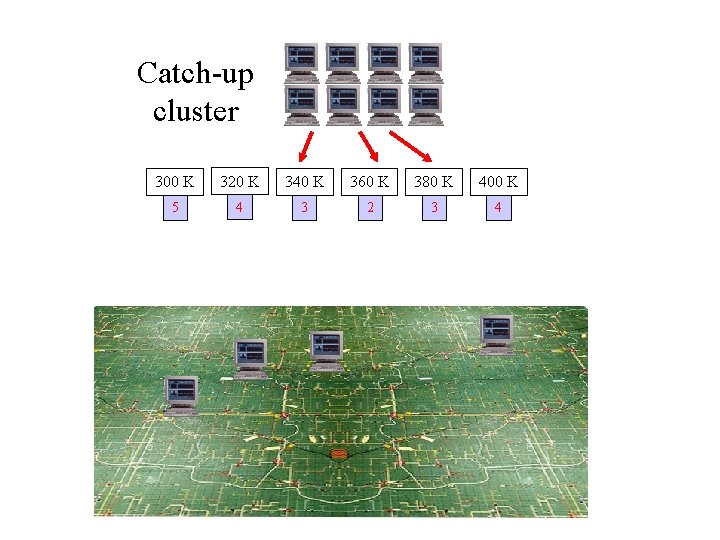
Catch-up cluster 300 K 320 K 340 K 360 K 380 K 400 K 5 4 3 2 3 4

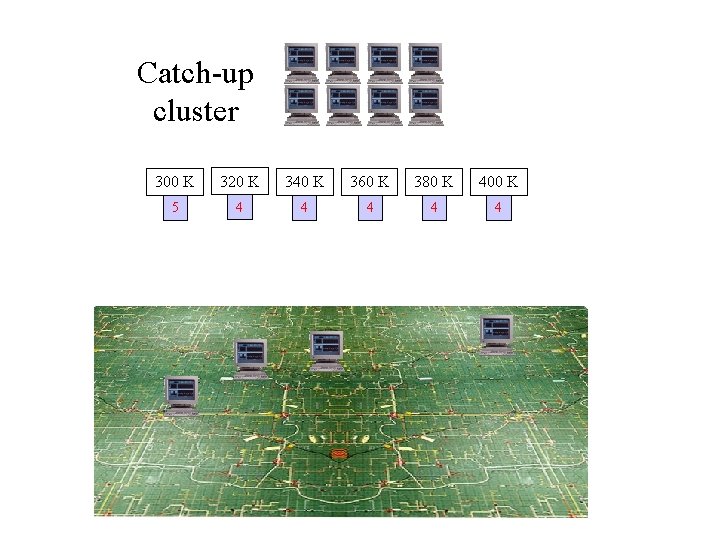
Catch-up cluster 300 K 320 K 340 K 360 K 380 K 400 K 5 4 4 4

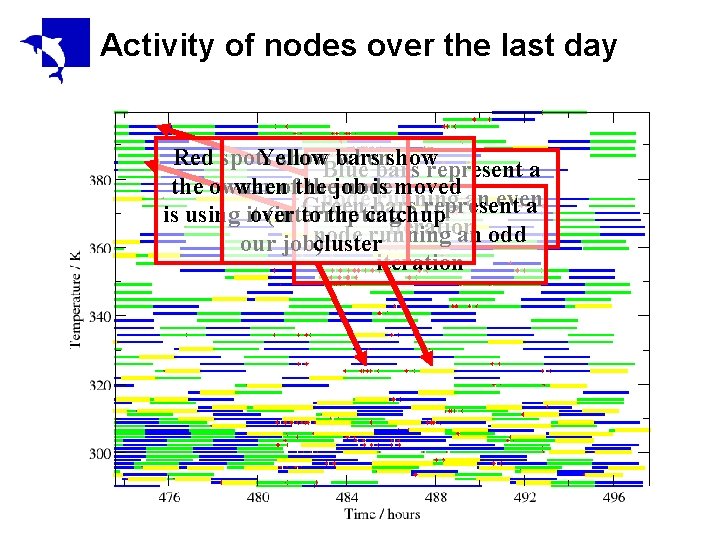
Activity of nodes over the last day Red spots Yellow show. Blue bars when showrepresent a bars the owner whenofthe job node isrunning moved an even node Green bars represent a is using itover (interrupting to the catchup iteration node running an odd our job)cluster iteration

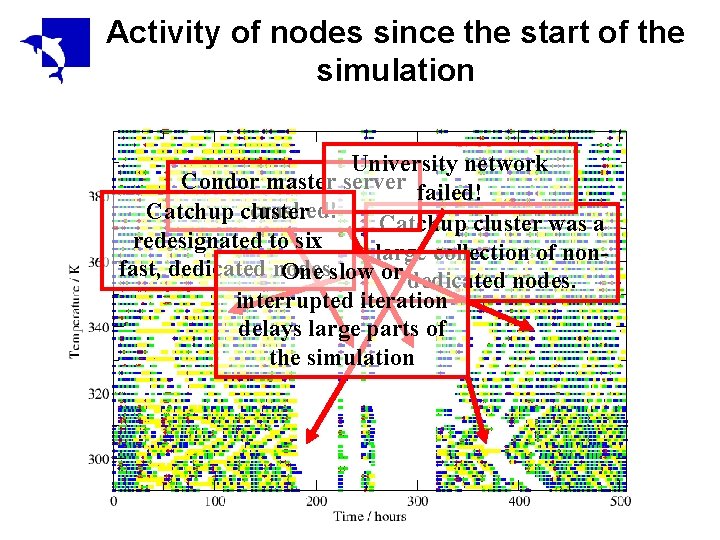
Activity of nodes since the start of the simulation University network Condor master server failed! crashed! Catchup cluster was a redesignated to six large collection of nonfast, dedicated nodes. One slow or dedicated nodes. interrupted iteration delays large parts of the simulation

Current paradigm for biomolecular simulation • Target selection: literature based; interesting protein/problem • System preparation: highly interactive, slow, idiosyncratic • Simulation: diversity of protocols • Analysis: highly interactive, slow, idiosyncratic • Dissemination: traditional – papers, talks, posters • Archival: archive data… and then lose the tape!

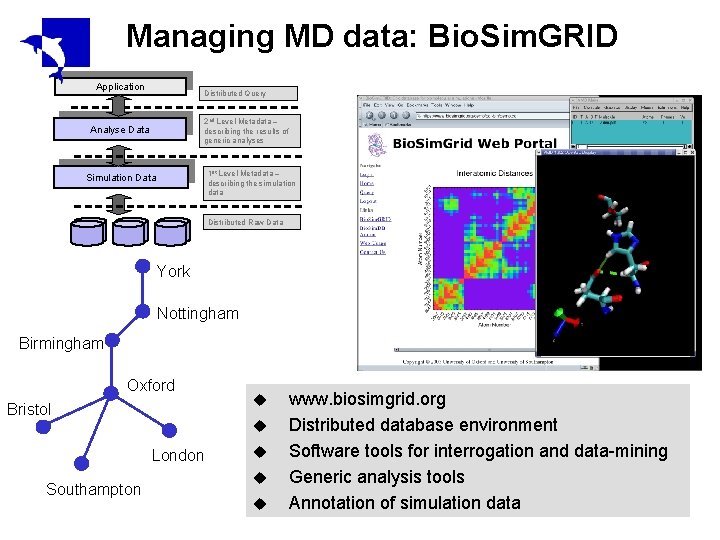
Managing MD data: Bio. Sim. GRID Application Distributed Query 2 nd Level Metadata – describing the results of generic analyses Analyse Data 1 st Level Metadata – describing the simulation data Simulation Data Distributed Raw Data York Nottingham Birmingham Oxford Bristol u London Southampton u u www. biosimgrid. org Distributed database environment Software tools for interrogation and data-mining Generic analysis tools Annotation of simulation data

Comparative simulations • Increase significance of results – Effect of force field – Simulation protocol • Long simulations and multiple simulations • Biology emerges from the comparisons – Very easy to over-interpret protein simulations – What’s noise, and what’s systematic?

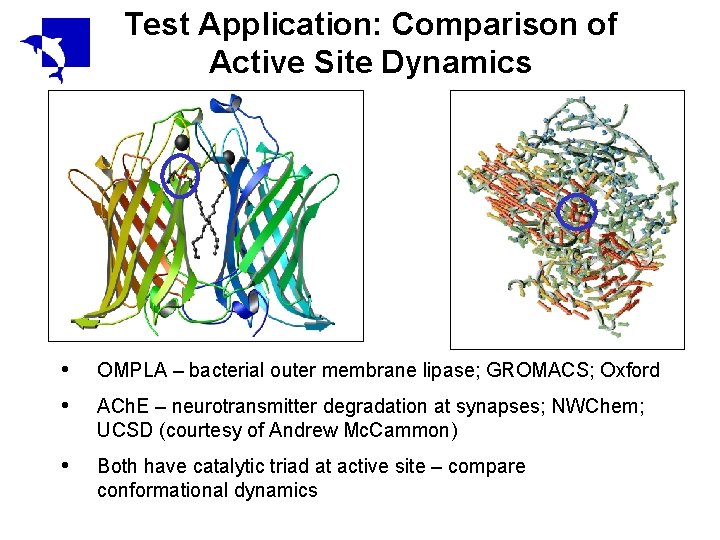
Test Application: Comparison of Active Site Dynamics • • OMPLA – bacterial outer membrane lipase; GROMACS; Oxford • Both have catalytic triad at active site – compare conformational dynamics ACh. E – neurotransmitter degradation at synapses; NWChem; UCSD (courtesy of Andrew Mc. Cammon)

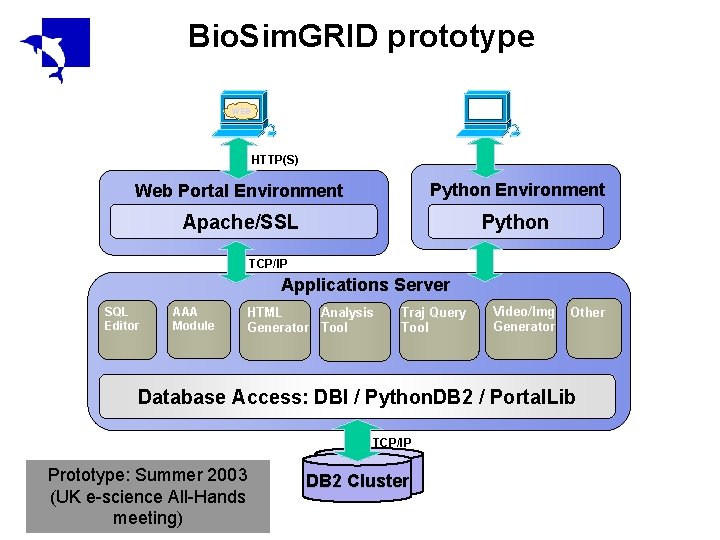
Bio. Sim. GRID prototype WEB HTTP(S) Web Portal Environment Python Environment Apache/SSL Python TCP/IP Applications Server SQL Editor AAA Module HTML Analysis Generator Tool Traj Query Tool Video/Img Generator Other Database Access: DBI / Python. DB 2 / Portal. Lib TCP/IP Prototype: Summer 2003 (UK e-science All-Hands meeting) DB 2 Cluster

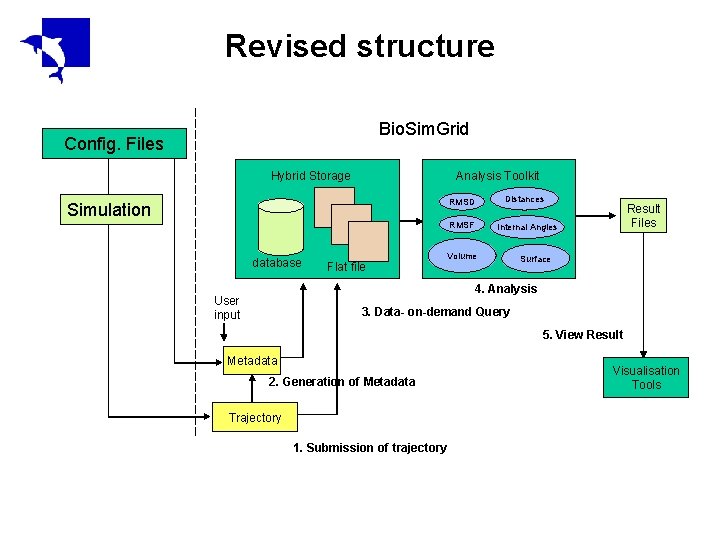
Revised structure Bio. Sim. Grid Config. Files Hybrid Storage Analysis Toolkit RMSD Simulation RMSF database Flat file Distances Result Files Internal Angles Volume Surface 4. Analysis User input 3. Data- on-demand Query 5. View Result Metadata 2. Generation of Metadata Trajectory 1. Submission of trajectory Visualisation Tools

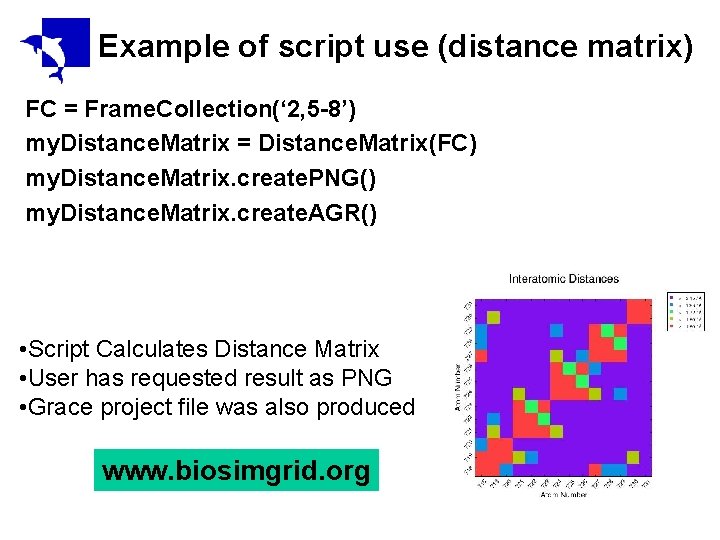
Example of script use (distance matrix) FC = Frame. Collection(‘ 2, 5 -8’) my. Distance. Matrix = Distance. Matrix(FC) my. Distance. Matrix. create. PNG() my. Distance. Matrix. create. AGR() • Script Calculates Distance Matrix • User has requested result as PNG • Grace project file was also produced www. biosimgrid. org

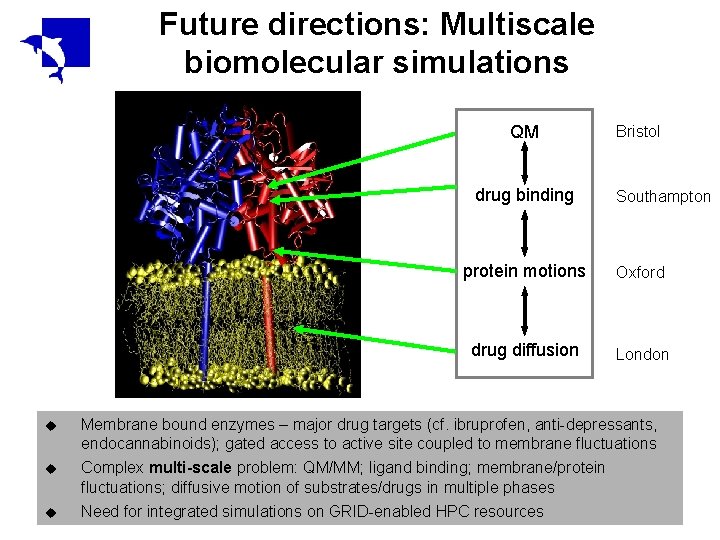
Future directions: Multiscale biomolecular simulations QM drug binding Bristol Southampton protein motions Oxford drug diffusion London u Membrane bound enzymes – major drug targets (cf. ibruprofen, anti-depressants, endocannabinoids); gated access to active site coupled to membrane fluctuations u Complex multi-scale problem: QM/MM; ligand binding; membrane/protein fluctuations; diffusive motion of substrates/drugs in multiple phases u Need for integrated simulations on GRID-enabled HPC resources

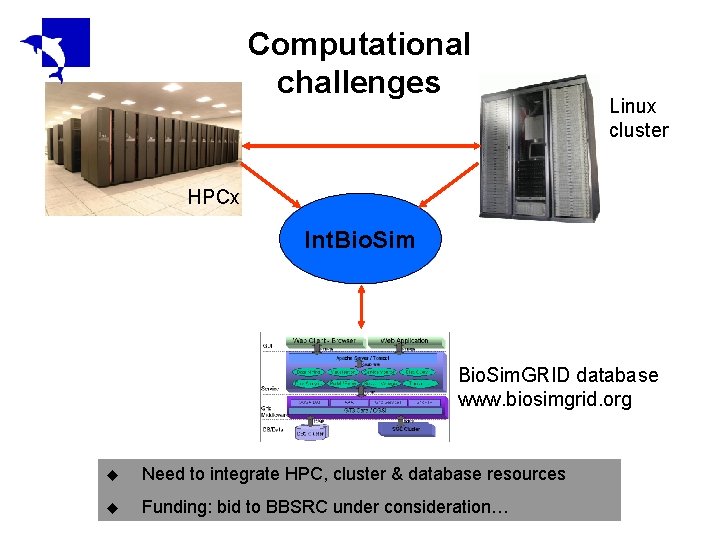
Computational challenges Linux cluster HPCx Int. Bio. Sim. GRID database www. biosimgrid. org u Need to integrate HPC, cluster & database resources u Funding: bid to BBSRC under consideration…

Acknowledgements My group: Stephen Phillips Richard Taylor Daniele Bemporad Christopher Woods Robert Gledhill Stuart Murdock My funding: BBSRC, EPSRC, Royal Society, Celltech, Astra. Zeneca, Aventis, Glaxo. Smith. Kline My collaborators: Mark Sansom, Adrian Mulholland, David Moss, Oliver Smart, Leo Caves, Charlie Laughton, Jeremy Frey, Peter Coveney, Hans Fangohr, Muan Hong Ng, Kaihsu Tai, Bing Wu, Steven Johnston, Mike King, Phil Jewsbury, Claude Luttmann, Colin Edge