Computational Radiology Laboratory Harvard Medical School www crl
Computational Radiology Laboratory Harvard Medical School www. crl. med. harvard. edu Brigham and Women’s Hospital Children’s Hospital Boston Massachusetts A validation framework for brain tumor segmentation Neculai Archip, Ph. D. Harvard Medical School
Outline • • • Brain image database; Existent segmentation data; STAPLE; How to validate a new algorithm; Performance study. Computational Radiology Laboratory. Slide 2
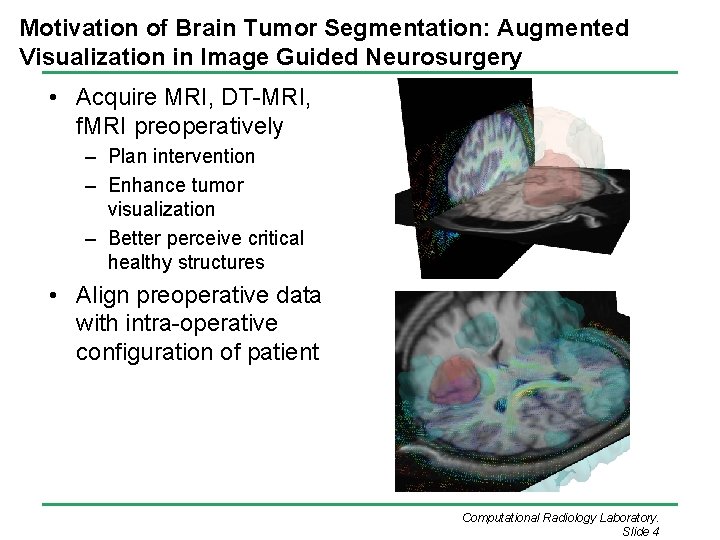
Motivation of Brain Tumor Segmentation: Augmented Visualization in Image Guided Neurosurgery • Acquire MRI, DT-MRI, f. MRI preoperatively – Plan intervention – Enhance tumor visualization – Better perceive critical healthy structures • Align preoperative data with intra-operative configuration of patient Computational Radiology Laboratory. Slide 4
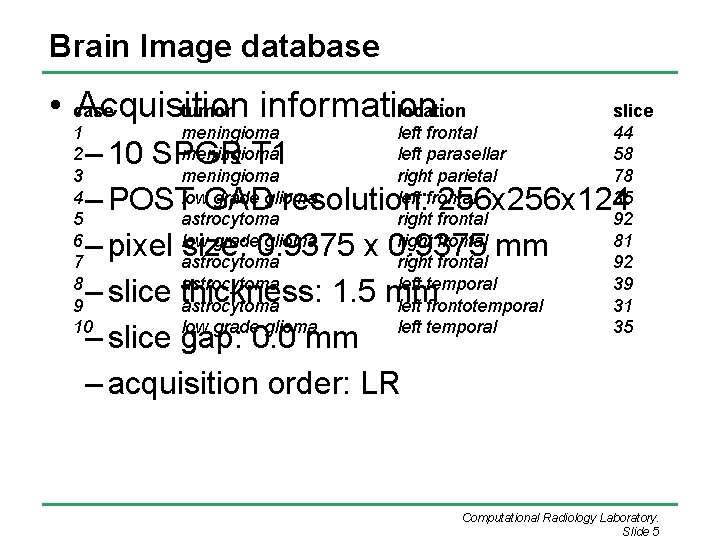
Brain Image database • case Acquisition information: tumor location 1 2 3 4 5 6 7 8 9 10 meningioma low grade glioma astrocytoma low grade glioma left frontal left parasellar right parietal left frontal right frontal left temporal left frontotemporal left temporal slice 44 58 78 35 92 81 92 39 31 35 – 10 SPGR T 1 – POST GAD resolution: 256 x 124 – pixel size: 0. 9375 x 0. 9375 mm – slice thickness: 1. 5 mm – slice gap: 0. 0 mm – acquisition order: LR Computational Radiology Laboratory. Slide 5
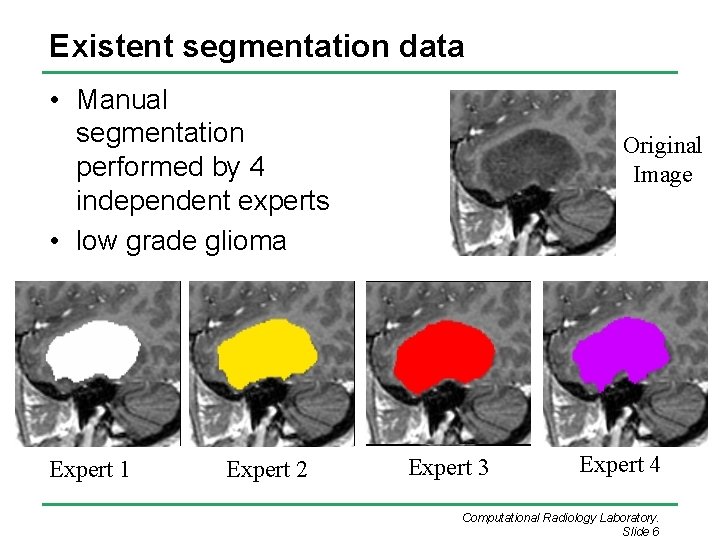
Existent segmentation data • Manual segmentation performed by 4 independent experts • low grade glioma Expert 1 Expert 2 Original Image Expert 3 Expert 4 Computational Radiology Laboratory. Slide 6
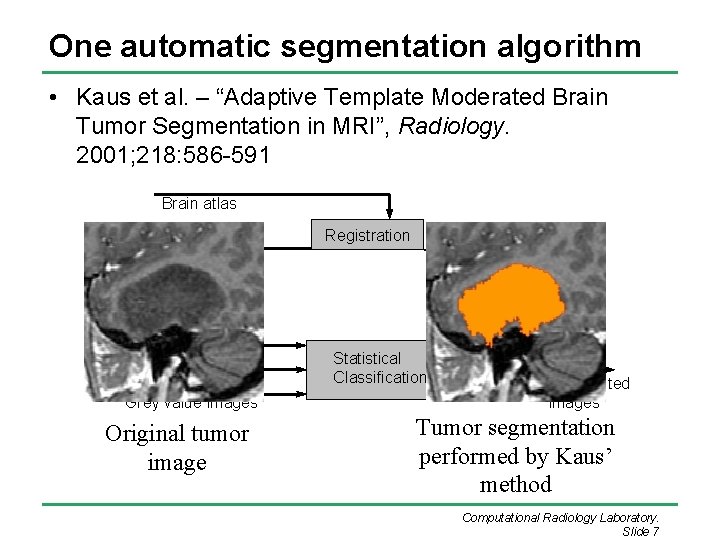
One automatic segmentation algorithm • Kaus et al. – “Adaptive Template Moderated Brain Tumor Segmentation in MRI”, Radiology. 2001; 218: 586 -591 Brain atlas Registration Template Distance Transforms Statistical Classification Grey value images Original tumor image Segmented images Tumor segmentation performed by Kaus’ method Computational Radiology Laboratory. Slide 7
Validation of Image Segmentation • Spectrum of accuracy versus realism in reference standard. • Digital phantoms. – Ground truth known accurately. – Not so realistic. • Acquisitions and careful segmentation. – Some uncertainty in ground truth. – More realistic. • Autopsy/histopathology. – Addresses pathology directly; resolution. • Clinical data ? – Hard to know ground truth. – Most realistic model. Computational Radiology Laboratory. Slide 8
Validation of Image Segmentation • Comparison to digital and physical phantoms: – Excellent for testing the anatomy, noise and artifact which is modeled. – Typically lacks range of normal or pathological variability encountered in practice. Computational Radiology Laboratory. Slide 9
Validation of Image Segmentation • Comparison to expert performance; to other algorithms: • What is the appropriate measure for such comparisons ? • Our new approach: • Simultaneous estimation of hidden ``ground truth’’ and expert performance. • Enables comparison between and to experts. • Can be easily applied to clinical data exhibiting range of normal and pathological variability. Computational Radiology Laboratory. Slide 11
STAPLE • STAPLE (Simultaneous Truth and Performance Level Estimation): – An algorithm for estimating performance and ground truth from a collection of independent segmentations. – Warfield, Zou, Wells MICCAI 2002. – Warfield, Zou, Wells, IEEE TMI 2004. Computational Radiology Laboratory. Slide 12
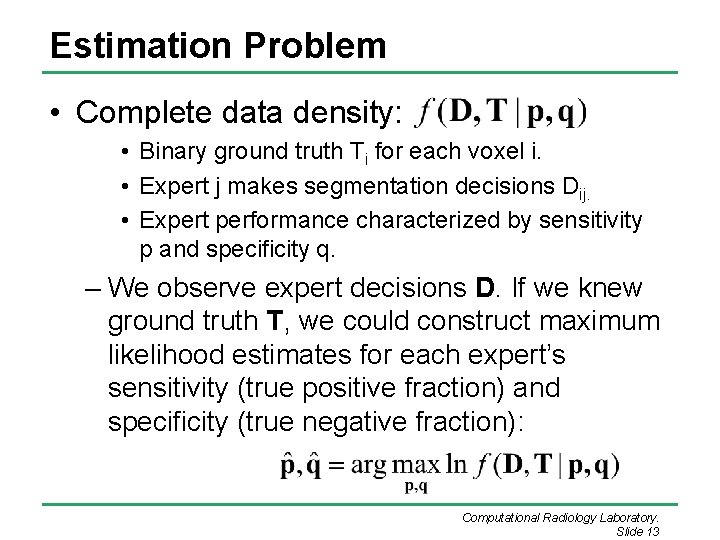
Estimation Problem • Complete data density: • Binary ground truth Ti for each voxel i. • Expert j makes segmentation decisions Dij. • Expert performance characterized by sensitivity p and specificity q. – We observe expert decisions D. If we knew ground truth T, we could construct maximum likelihood estimates for each expert’s sensitivity (true positive fraction) and specificity (true negative fraction): Computational Radiology Laboratory. Slide 13
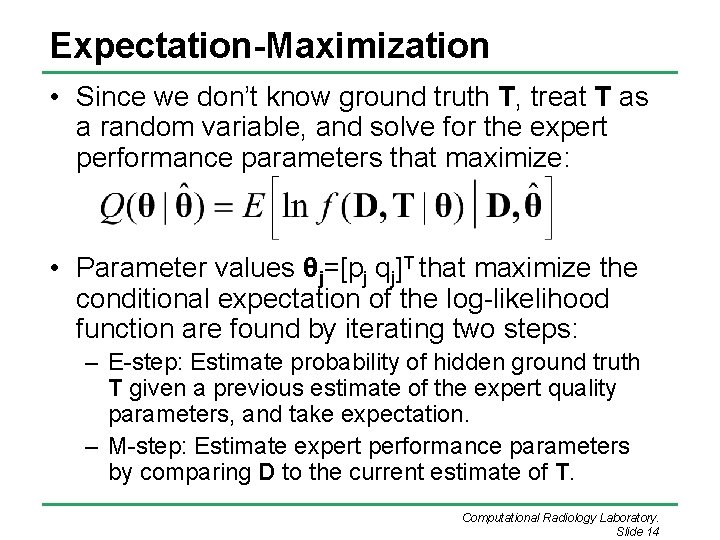
Expectation-Maximization • Since we don’t know ground truth T, treat T as a random variable, and solve for the expert performance parameters that maximize: • Parameter values θj=[pj qj]T that maximize the conditional expectation of the log-likelihood function are found by iterating two steps: – E-step: Estimate probability of hidden ground truth T given a previous estimate of the expert quality parameters, and take expectation. – M-step: Estimate expert performance parameters by comparing D to the current estimate of T. Computational Radiology Laboratory. Slide 14
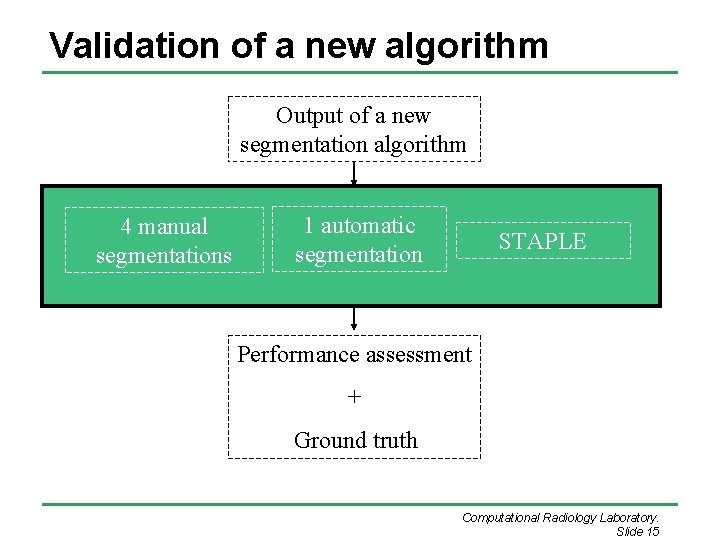
Validation of a new algorithm Output of a new segmentation algorithm 4 manual segmentations 1 automatic segmentation STAPLE Performance assessment + Ground truth Computational Radiology Laboratory. Slide 15
A new algorithm • Spectral clustering algorithms: – Shi and Malik 2000 • NCUT criterion – Ng, Jordan and Weiss 2002 • Supervised clustering using k eigenvectors – Miela and Shi 2002 • Supervised clustering – connection with Markov Chains – Fowlkes, Belongie, Chung, Malik 2004 • Nyström method – spine segmentation from MRI • Fiedler eigenvector based segmentation: – Archip et al. 2005. • Related approached used in seriation and the consecutive ones problems. Computational Radiology Laboratory. Slide 16
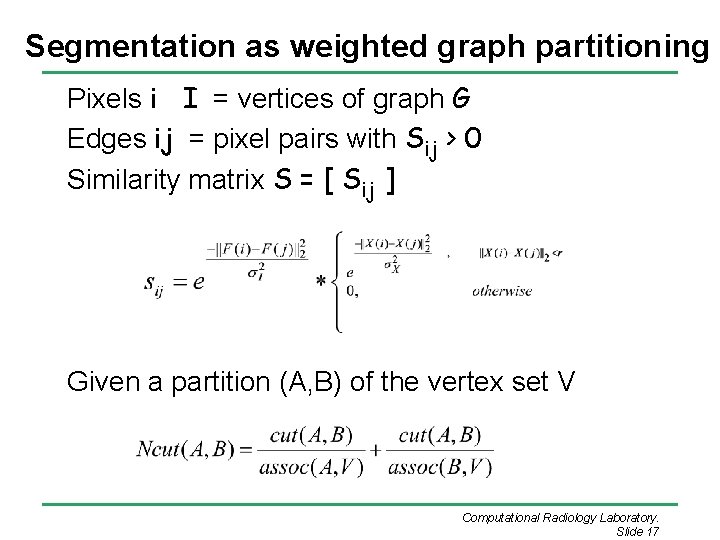
Segmentation as weighted graph partitioning Pixels i I = vertices of graph G Edges ij = pixel pairs with Sij > 0 Similarity matrix S = [ Sij ] Given a partition (A, B) of the vertex set V Computational Radiology Laboratory. Slide 17
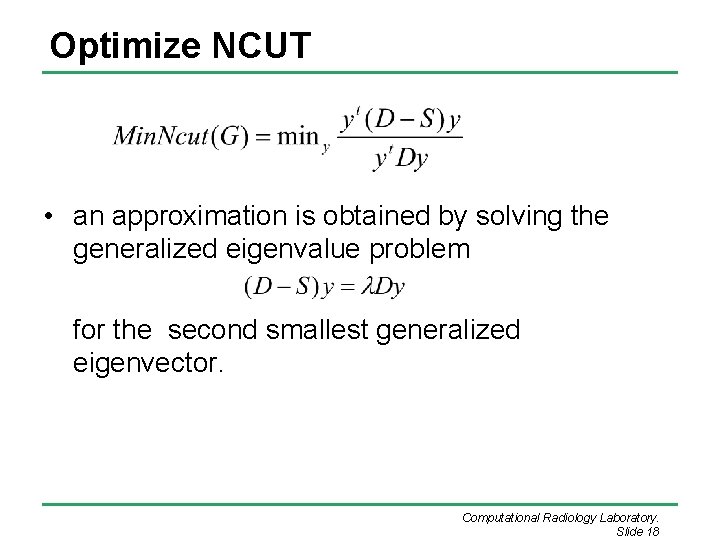
Optimize NCUT • an approximation is obtained by solving the generalized eigenvalue problem for the second smallest generalized eigenvector. Computational Radiology Laboratory. Slide 18
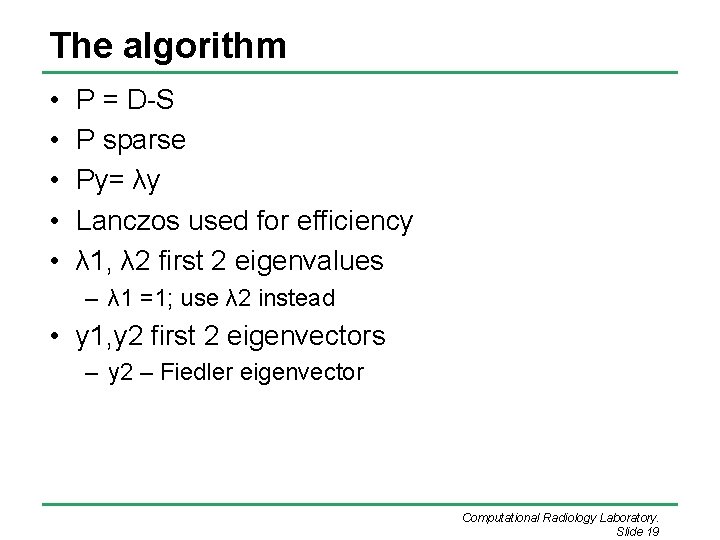
The algorithm • • • P = D-S P sparse Py= λy Lanczos used for efficiency λ 1, λ 2 first 2 eigenvalues – λ 1 =1; use λ 2 instead • y 1, y 2 first 2 eigenvectors – y 2 – Fiedler eigenvector Computational Radiology Laboratory. Slide 19
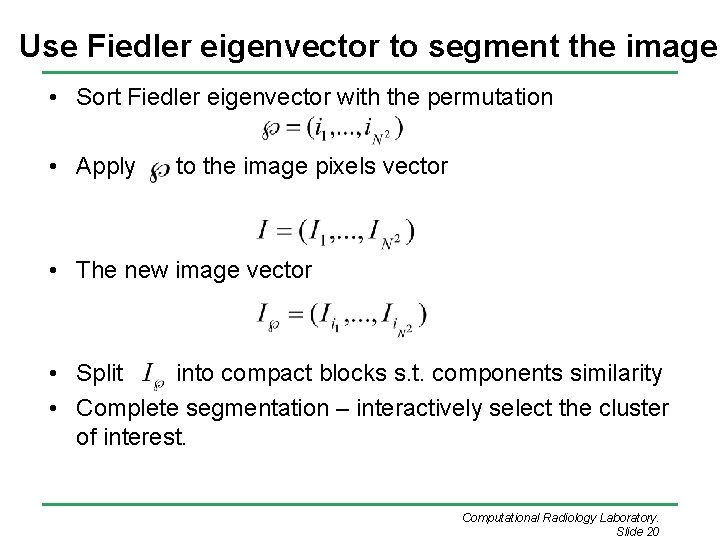
Use Fiedler eigenvector to segment the image • Sort Fiedler eigenvector with the permutation • Apply to the image pixels vector • The new image vector • Split into compact blocks s. t. components similarity • Complete segmentation – interactively select the cluster of interest. Computational Radiology Laboratory. Slide 20
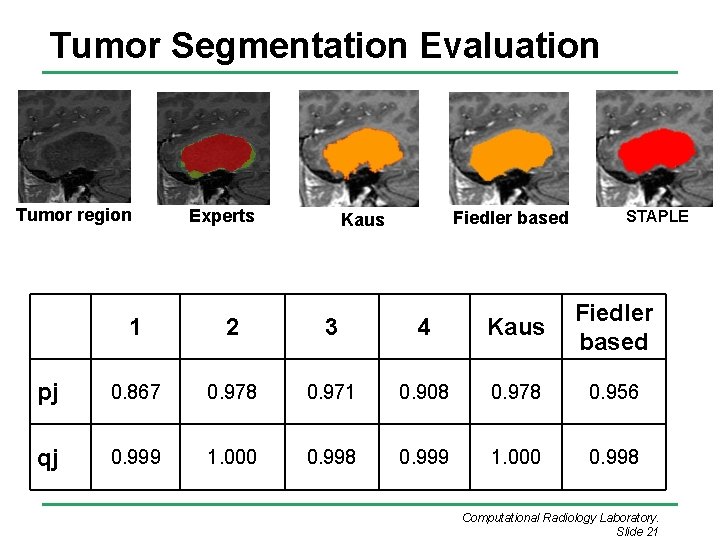
Tumor Segmentation Evaluation Tumor region Experts Fiedler based Kaus STAPLE 1 2 3 4 Kaus Fiedler based pj 0. 867 0. 978 0. 971 0. 908 0. 978 0. 956 qj 0. 999 1. 000 0. 998 Computational Radiology Laboratory. Slide 21
Conclusions • Framework for the validation of brain tumor segmentation: image + software. • STAPLE public available. • Image and segmentation data will be made public available. • Existent data to be added to the image database. Computational Radiology Laboratory. Slide 22

Acknowledgements Contributors to this research: • • • Simon K. Warfield. Peter M. Black. Alexandra Golby. Ferenc A. Jolesz. Ron Kikinis. Lawrence Panych. Kelly H. Zou. Steve Haker. Vicente Grau-Colomer. Olivier Clatz Herve Delingette • • • Herve Delingette Nicholas Ayache Martha Shenton. Clare Tempany. Carl Winalski. Michael Kaus. William M. Wells. Andrea Mewes. Heidelise Als. Petra Huppi. Terrie Inder. www. crl. med. harvard. edu Computational Radiology Laboratory. Slide 23
- Slides: 21