Complex trait analysis development and genomics The Complex









- Slides: 28

Complex trait analysis, development, and genomics The Complex Trait Consortium and the Collaborative Cross Rob Williams, Gary Churchill, and members of the Complex Trait

Material included in handouts and on the CD also see www. complextrait. org “Solving the puzzle of complex diseases, from obesity to cancer, will require a holistic understanding of the interplay between factors such as genetics, diet, infectious agents, environment, behavior, and social Elias Zerhouni: The NIH Roadmap. Science 302: 63 structures. ” (2003)

A group of ~150 mouse geneticists most of whom have interests in pervasive diseases and differences in disease susceptibility. Main catalysts and models ENU mutagenesis programs Sequencing and SNP consortia General Aim: Improve resources for complex trait analysis using mice. • Catalyze genotyping of strains • Simulation studies of crosses • Planning a collaborative cross • Improved use of resources

The short chronology of the CTC 1. Established Nov 2001, Edinburgh (n = 20) 2. 1 st CTC Conference, May 2002, Memphis (n = 80; hosted by R Williams) 3. CTC Collaborative Cross design workshop, Aug 2002, JHU (K Broman and R Reeves, host) 4. CTC Satellite meeting at IMGC Nov 2003 (n = 40) 5. 2 nd CTC Conference, July 2003, Oxford (n = 80; hosted by R Mott and J Flint) 6. CTC strain selection workshop, Sept 2003 (M Daly, host) 7. 3 rd CTC Conference, July 2004, TJL

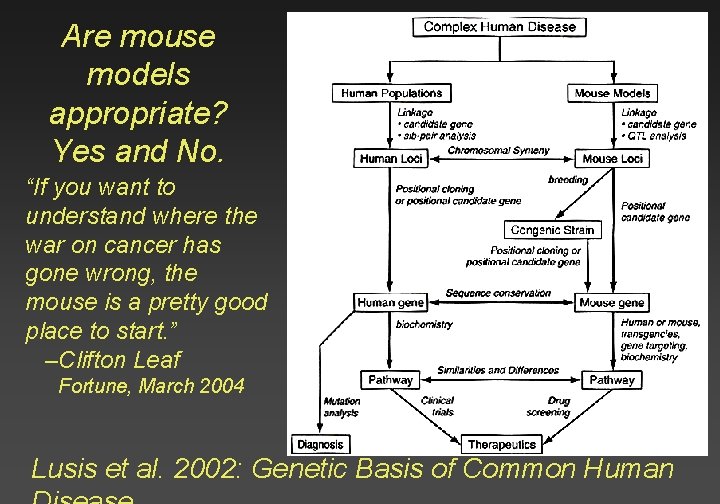
Are mouse models appropriate? Yes and No. “If you want to understand where the war on cancer has gone wrong, the mouse is a pretty good place to start. ” –Clifton Leaf Fortune, March 2004 Lusis et al. 2002: Genetic Basis of Common Human

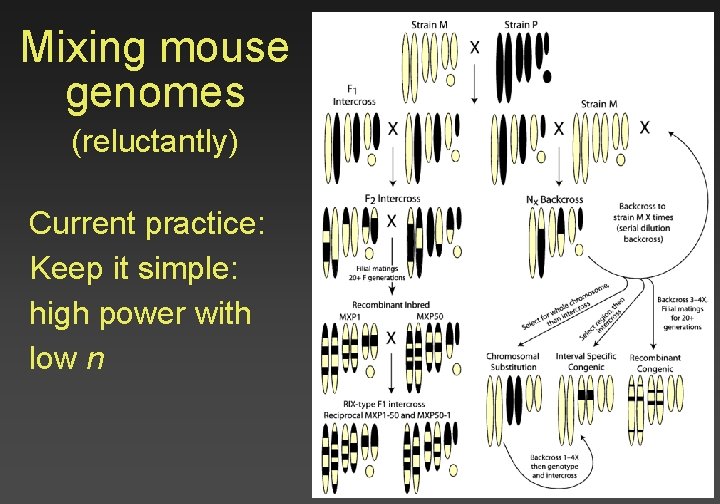
Mixing mouse genomes (reluctantly) Current practice: Keep it simple: high power with low n

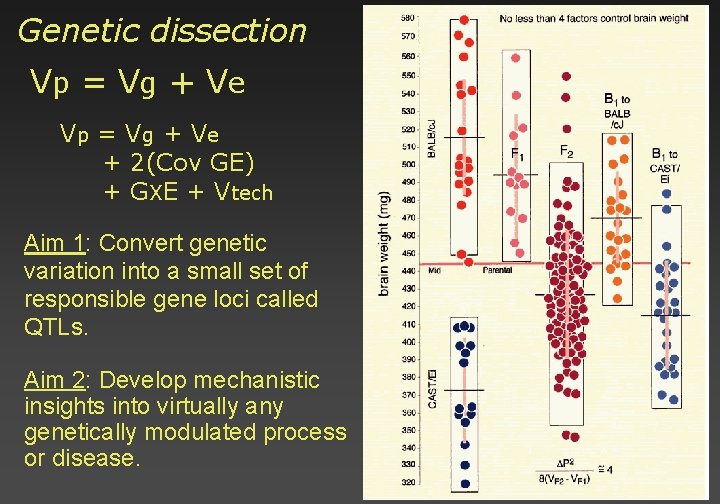
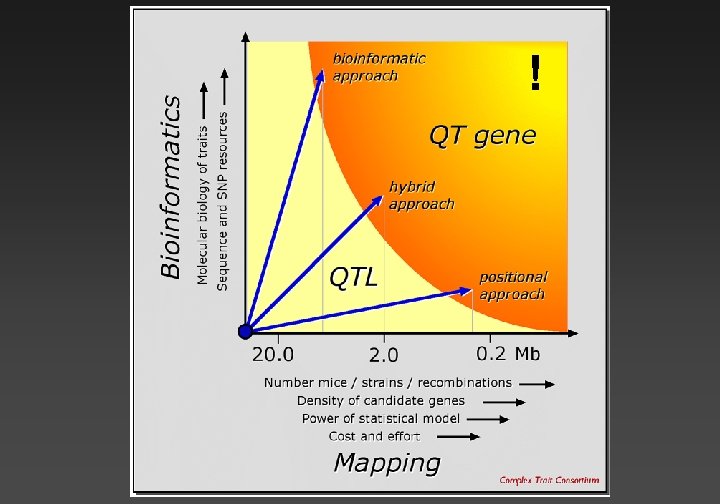
Genetic dissection Vp = V g + V e + 2(Cov GE) + GXE + Vtech Aim 1: Convert genetic variation into a small set of responsible gene loci called QTLs. Aim 2: Develop mechanistic insights into virtually any genetically modulated process or disease.

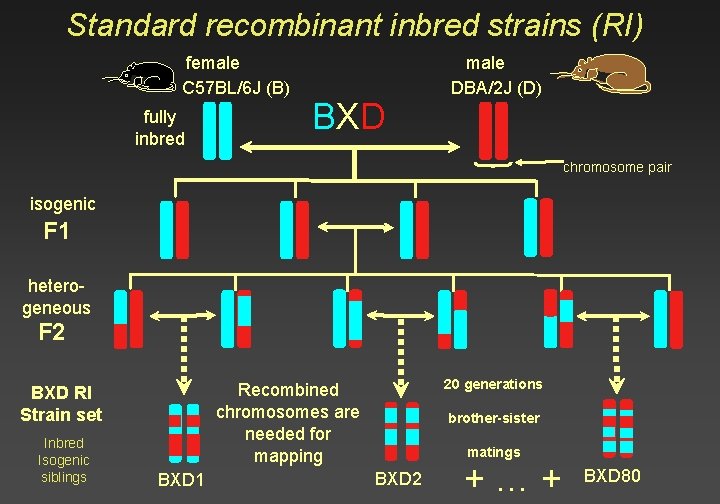
Standard recombinant inbred strains (RI) female C 57 BL/6 J (B) fully inbred BXD male DBA/2 J (D) chromosome pair isogenic F 1 heterogeneous F 2 Inbred Isogenic siblings 20 generations Recombined chromosomes are needed for mapping BXD RI Strain set BXD 1 brother-sister matings BXD 2 +…+ BXD 80

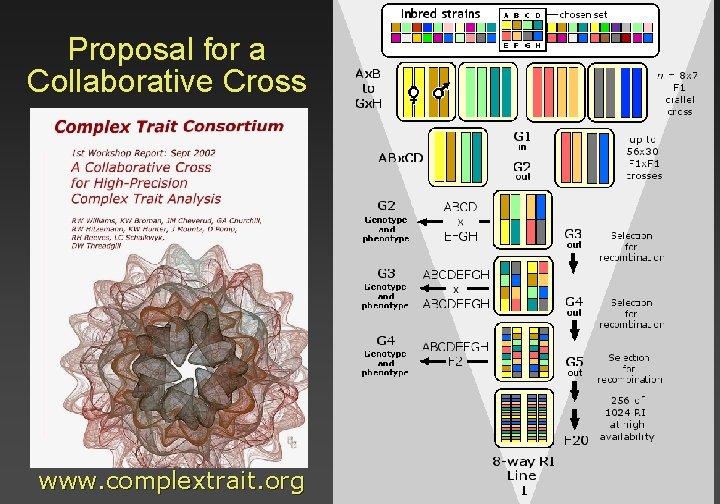
Proposal for a Collaborative Cross www. complextrait. org

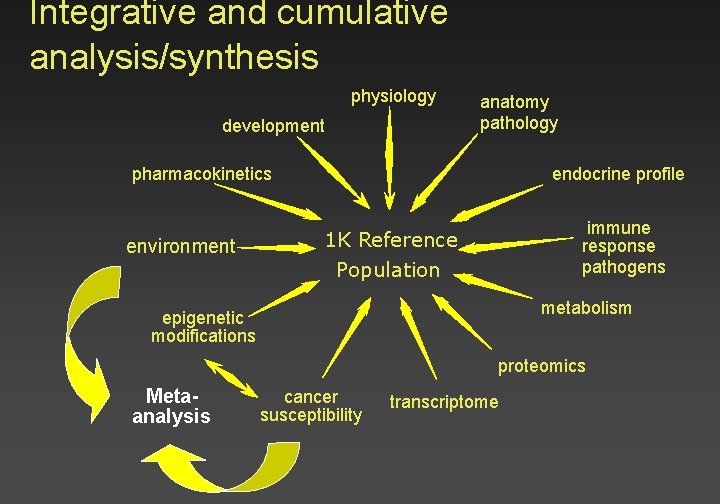
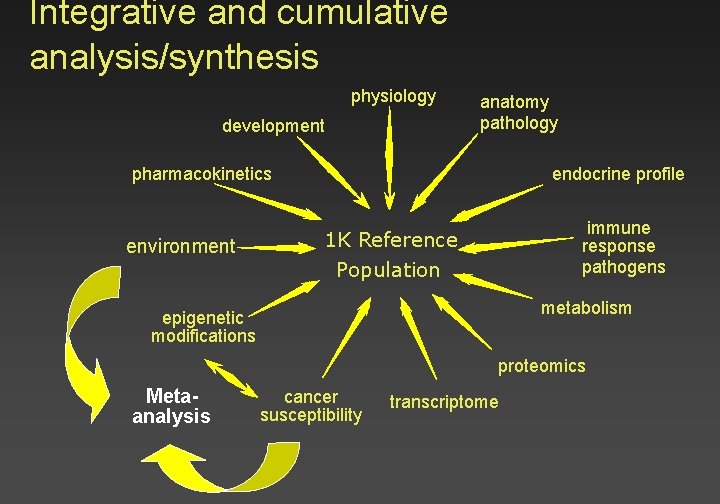
Integrative and cumulative analysis/synthesis physiology development anatomy pathology endocrine profile pharmacokinetics environment immune response pathogens 1 K Reference Population metabolism epigenetic modifications proteomics Metaanalysis cancer susceptibility transcriptome

Design criteria for a Collaborative Cross Broad utility: a resource that combines diverse haplotypes and that harbors a broad spectrum of alleles Freedom from genotyping. Lowering the entry barrier into this field Unrestricted access to strains, tissues, data, and statistical analysis suites (on-line mapping) Improved power and precision for trait mapping. Epistasis! Powerful new approaches to analysis of complex systems. Pleiotropy Analysis of gene-by-environment interactions A systems biology resource A new type of complex animal model to study common human diseases

A set of 420 RI lines

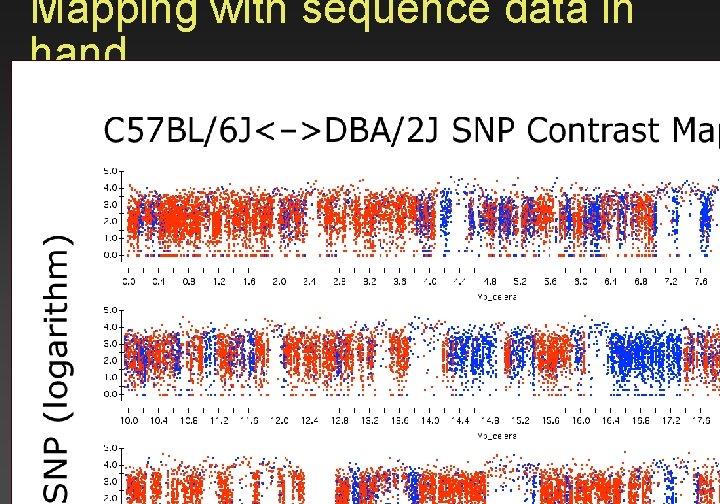
Mapping with sequence data in hand

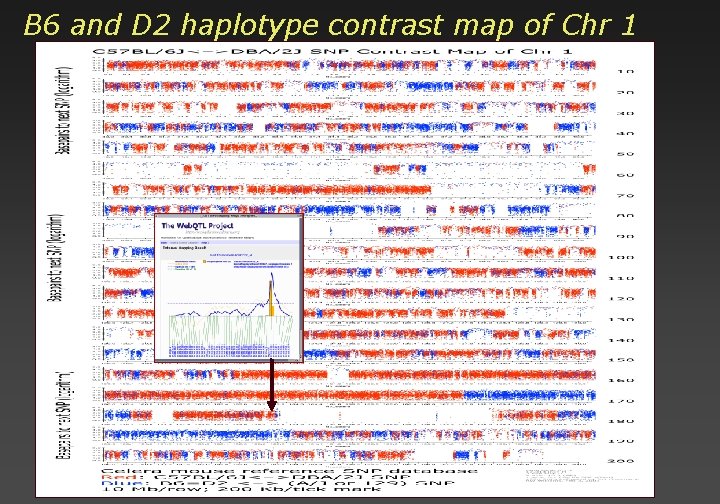
B 6 and D 2 haplotype contrast map of Chr 1

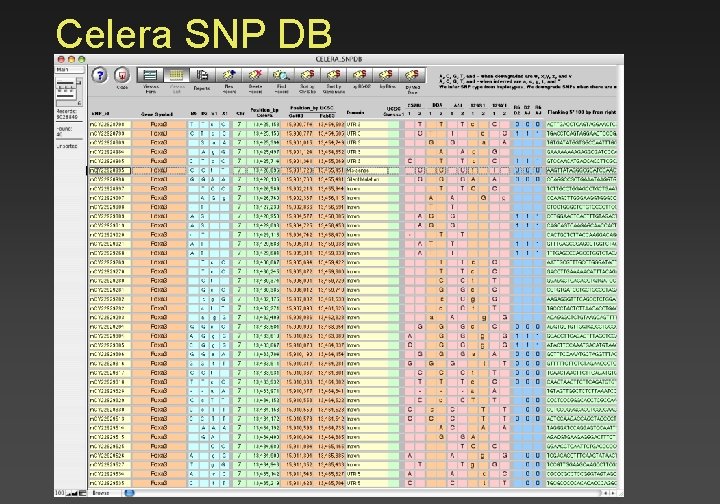
Celera SNP DB


Integrative and cumulative analysis/synthesis physiology development anatomy pathology endocrine profile pharmacokinetics environment immune response pathogens 1 K Reference Population metabolism epigenetic modifications proteomics Metaanalysis cancer susceptibility transcriptome

Wilt Chamberlain: 7 feet 1 inch Willie Shoemaker: 4 feet 11 inches 1. 44 -fold Phenotypes: from highly complex such as body size to highly specific, such as transcript expression difference 6 6 QTL/QT gene 24 th www. webqtl. org

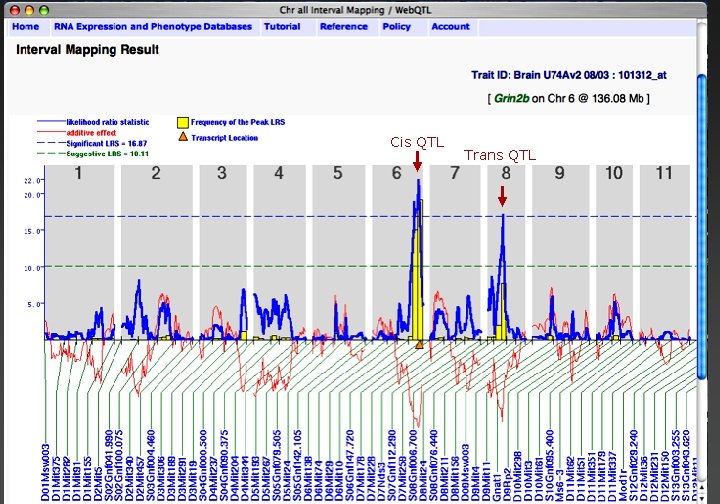
Grin 2 b Cis QTL Trans QTL

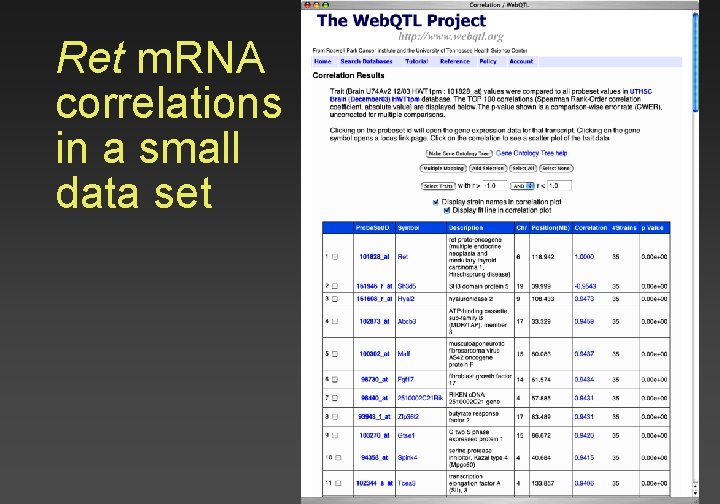
Ret m. RNA correlations in a small data set

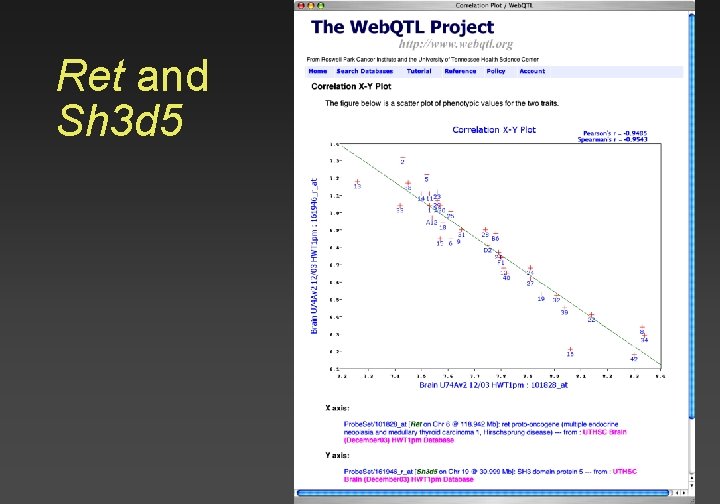
Ret and Sh 3 d 5

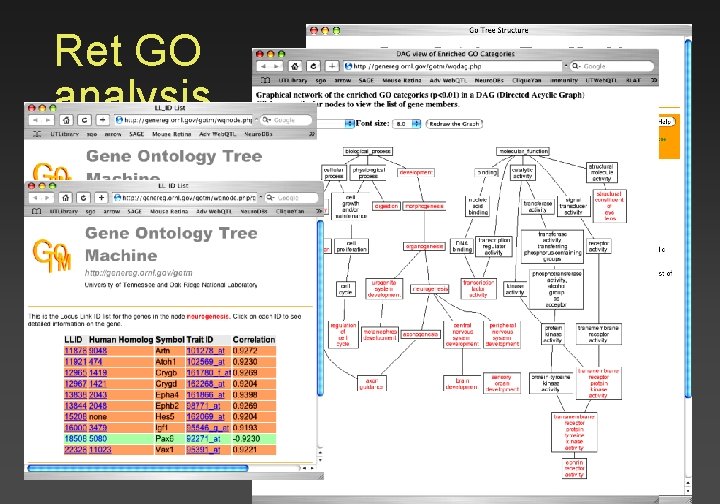
Ret GO analysis

Handdrawn sketch of the App neighborhood

Associational Networks QTL networks add layer of shared causality

Integrative and cumulative analysis/synthesis physiology development anatomy pathology endocrine profile pharmacokinetics environment immune response pathogens 1 K Reference Population metabolism epigenetic modifications proteomics Metaanalysis cancer susceptibility transcriptome

Cost Components: 24– 28 M over 7– 8 yrs Per diem for 8, 000 to 10, 000 cages (~1500 K/year) Genotyping intermediate generations (~500 K/year) Prospective tissue harvesting and cryopreservation (~500 K/year) Molecular phenotyping of select tissue as proof-ofprinciple (500 K/year) Bioinformatics, statistical modeling, administration, colony management (~500 K/year) Cryopreservation of final lines at F 25+ (~200 K) Sequencing of parental strains (unfunded)


Collaborators Lu Lu Elissa Chesler David Airey Siming Shou Jing Gu Yanhua Qu Ken Manly (UTHSC) David Threadgill (UNC Chapel Hill) Bob Hitzemann (OHSU) Gary Churchill (TJL) Fernando Pardo Manuel de Villena (UNC) Karl Broman (JHU) Dan Gaile (SUNY Buffalo) Kent Hunter (NCI) Jay Snoddy (ORNL) Jim Cheverud (Wash U) Tim Wiltshire (GNF) Supported by: NIAAA-INIA Program, NIMH, NIDA, and the National Science Foundation (P 20 -MH 62009), NEI, a Human Brain Project and the William and Dorothy Dunavant Endowment.