Complementary DNA Sequencing Expressed Sequence Tags and Human





- Slides: 18

Complementary DNA Sequencing: Expressed Sequence Tags and Human Genome Project Adams MD, Kelly JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, Mc. Combie WR, and Venter JC* Presented by Malva “Lisa” Severios

Background • Partial DNA sequencing done on 600 human c. DNA clones to generate expressed sequence tags (ESTs). • With up to 100, 000 genes present in the human genome, 30, 000 are expected to be expressed in the brain.

Expressed Sequence Tags • Markers for Genome mapping. • Obtained by randomly acquiring c. DNA sequences. • Quick method that’s able to give information about the diversity of genes expressed. • Can be mapped to chromosomes by FISH, RH panels, BAC contigs, and Polymorphic STS. • Uses include: determining relationships between genes and forensic analysis.

Purpose • Promote a faster approach to c. DNA characterization. • Tag most of the human neurological genes expressed in the brain. • Up to 1/4 th of all genetic diseases affect neurological functions, indicating the importance of the genes expressed in the brain. • Provide a less expensive method to find human genes.

Purpose…cont. • Examine diversity of representative c. DNA libraries. • Identify advantages / disadvantages of c. DNA libraries. • Determine info content and accuracy of single-run sequence reactions.

Experimental Goal Of c. DNA libraries, random-primed and partial c. DNA clones are more informative in identifying genes and constructing a more useful EST database than sequencing from the ends of fulllength c. DNAs. Therefore, obtain coding sequences in order to take advantage of more sensitive peptide sequences and for nucleotide sequence comparisons.

Experiment • 3 commercial human brain c. DNA libraries ~ made from isolated m. RNA in the hippocampus and temoral cortex of a 2 year old and fetus~ • Single-run DNA sequence data obtained from randomly chosen c. DNA clones. • Tested subtractive hybridization to increase the # of brain-specific clones.

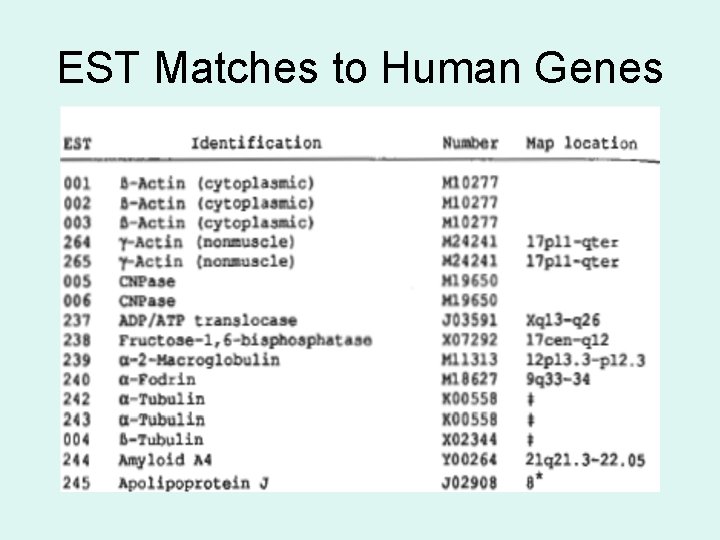
EST Matches to Human Genes

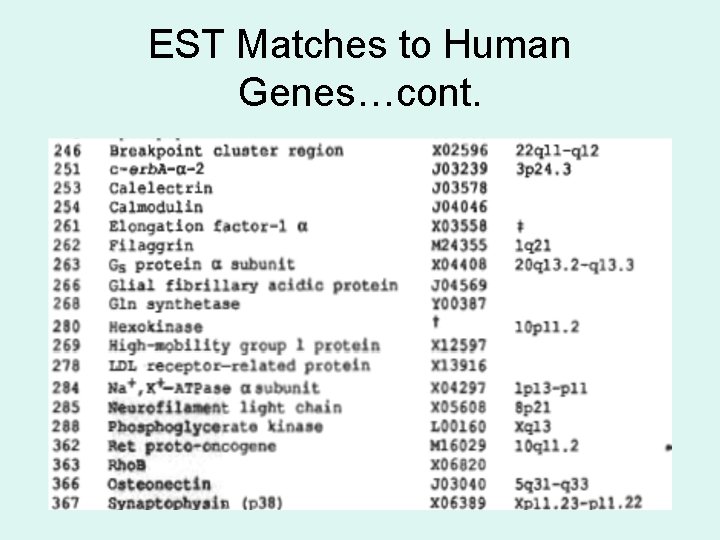
EST Matches to Human Genes…cont.

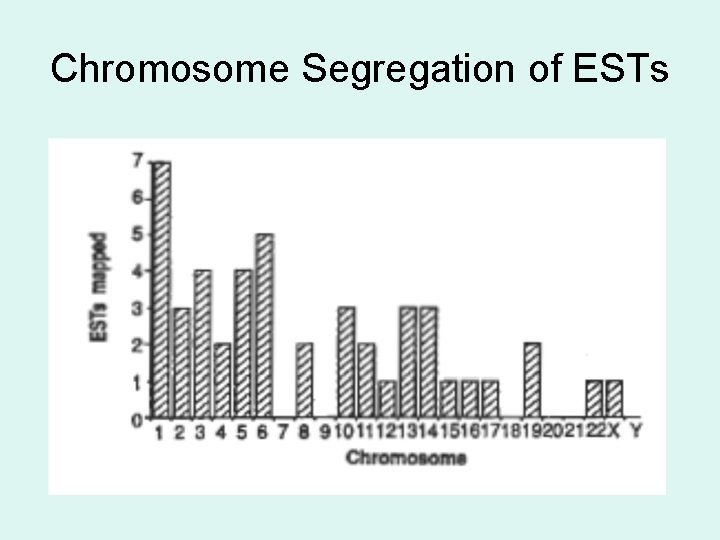
Map ESTs to Human Chromosomes • PCR used to screen somatic cell hybrid cell lines with defined sets of human chromosomes → hybrids with human gene corresponding to a specific EST yields an amplified fragment. • The chromosome present in all hybrids that yielded an amplified fragment is the location of the EST.

Chromosome Segregation of ESTs

Gen. Bank • Able to analyze accuracy of ds automated DNA sequencing by matching ESTs with human sequences in Gen. Bank. • NIH genetic sequence database, providing a collection of DNA sequences to the public. • Avg accuracy rate: 97. 7% for 150 - 400 bases, having < 3% ambiguous base calls. • 348 ESTs met this criteria and were submitted to Gen. Bank.

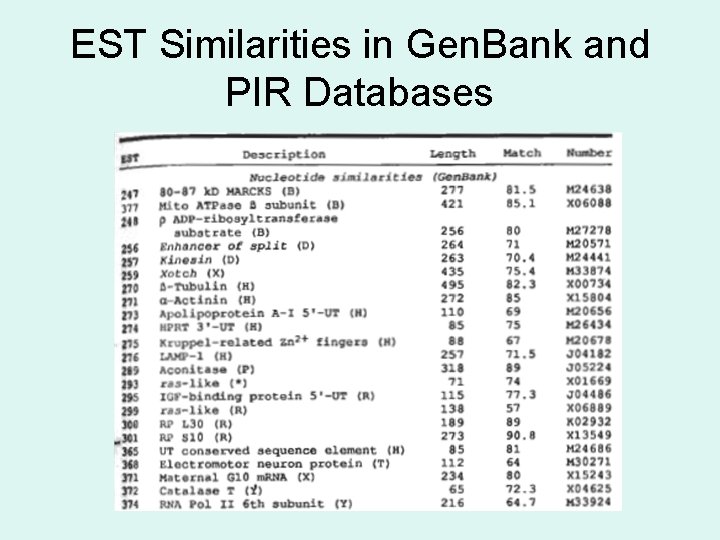
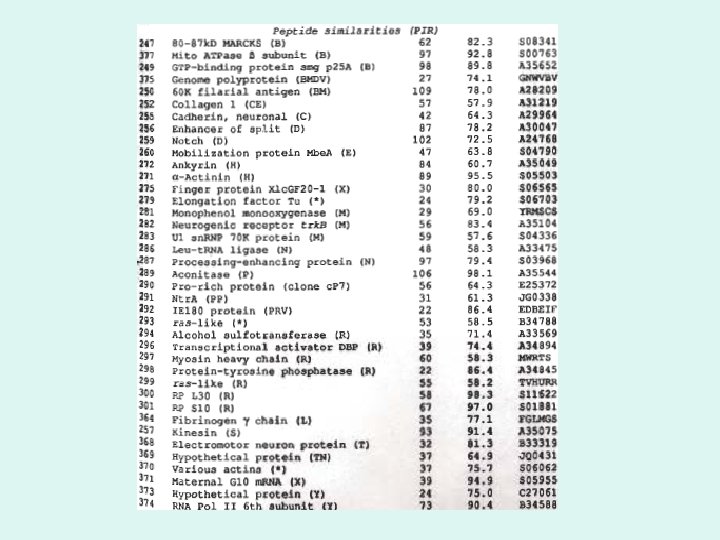
EST Similarities in Gen. Bank and PIR Databases


Conclusions • Single-run DNA sequencing is efficient in obtaining preliminary data on c. DNA clones. • Found 230 ESTs, representing new genes • Random selection approach yields a high amount of highly represented clones in the c. DNA libraries used ~ NOT GOOD!! • EST and physical mapping → high resolution map of the location of genes on chromosomes ~ more efficient and cheaper than genomic sequencing.

Conclusions…cont. • Using ESTs will provide a better way of anayzing chromosomes and discovering more human genes. • EST method will result in partial sequencing of most human brain c. DNAs in a couple years → further identification of genes involved in neurological diseases.

What’s next? • Characterize the new 230 genes found by: ~ complete sequencing and expression ~ chromosome mapping ~ tissue distribution ~ immunology

Debate against c. DNA for the Human Genome Project • With c. DNAs, can only find sequence of protein encoding information (ie. Know sequence of exons, not introns ~ which is important for regulation and control). • Difficulty finding every single m. RNA expressed in all tissues, cell types, and developmental stages. • Gene coding regions, and therefore m. RNA sequences, are NOT predictable from genomic sequences → don’t need large-scale c. DNA sequencing.