Comparative Tools SRI International Bioinformatics Comparative Genomics Tools

Comparative Tools

SRI International Bioinformatics Comparative Genomics Tools l Several ways to compare PGDB data l All relevant PGDBs must be loaded into one instance of Pathway Tools l Compare objects across databases l Metabolic map comparisons l Compare genome regions around orthologous genes l Web-based comparative analysis tables
Second Database Selector SRI International Bioinformatics There are two different organism selectors in Bio. Cyc The first: found at the top of the page, selects a single organism/database l l. The second: a multi-organism selector, used for comparative analyses l Tools → Comparative analysis ; Choose Organisms button l On gene page: Genes → Select organisms/databases for comparisons
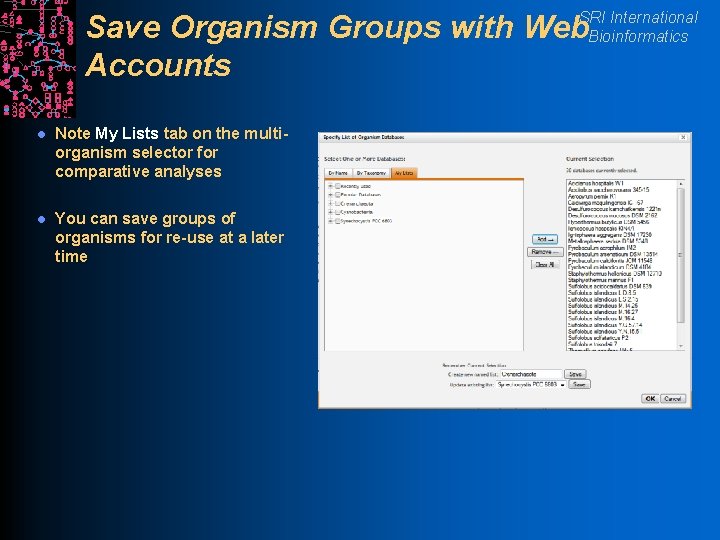
SRI International Bioinformatics Save Organism Groups with Web Accounts l Note My Lists tab on the multiorganism selector for comparative analyses l You can save groups of organisms for re-use at a later time
Compare Objects Across Databases l SRI International Bioinformatics Web mode l Find object in one other database u u l Gene -> Show this gene in another database [Type] -> Show this [type] in another database Find object in multiple other databases u u [Type] -> Select organisms/databases for comparison operations [Type] -> Show this [type] in multiple databases l Desktop mode l Right-click Show -> Frame in other DB l What does it mean for two objects to be the same? l Genes: same name or ortholog l Proteins: same name l Reactions, Pathways, Compounds: same object ID

Pathway Comparison SRI International Bioinformatics l Use Species Comparison button on pathway pages l Is the pathway inferred to be present? Evidence glyph l Green – reaction has an associated enzyme in this organism l Blue – the Hole Filler has predicted an enzyme for this reaction l Black – no enzyme, sorry l Orange – within this PGDB, this reaction is only assigned to this one pathway l Magenta – spontaneous reaction, or the mysterious “other” l l Enzymes and associated genes list l Grouped by reaction l Operons
Compare Metabolic Maps Via Cellular Overview SRI International Bioinformatics l Overviews -> Highlight -> Species Comparison l Desktop mode only l. A reaction is shared between two PGDBs if, for both PGDBs: l The reaction occurs spontaneously, or l An enzyme is present that catalyzes the reaction l If the same reaction exists as a pathway hole in two PGDBs, it will not be highlighted

SRI International Bioinformatics Compare Genome Regions l See a gene and its orthologs in their respective contexts l Genes coding for pathway enzymes l And how those genes are organized l Gene page: l Gene → Align in Multi-Genome Browser

SRI International Bioinformatics Web-based Comparative Analyses l Generate tables comparing the presence of biological entities in different organisms l Select what you want to compare l Select the organisms you want to compare l Will generate a table with items you can click through on
Comparative Analysis Example l Select SRI International Bioinformatics Pathways and three organisms l Multiple tables result l Pathways by class l Presence of pathway holes l Each type of item (pathways, reactions, etc) has different tables l Clicking on a pathway class generates a table comparing the actual pathways within that class l Can then click through to cross-species comparison (like from the pathway pages)
- Slides: 10