Comparative Proteomics Protein Profiling o Understanding the basics
Comparative Proteomics Protein Profiling o. Understanding the basics o. SDS Electrophoresis Physics & Chemistry

Can biomolecular evidence be used to determine evolutionary relationships? o o Traits are the result of: n Structure n Function Proteins determine structure and function DNA codes for proteins that confer traits Changes in DNA lead to proteins with: n n n o Different functions Novel traits Positive, negative, or no effects Genetic diversity provides pool for natural selection = evolution

Proteomics o Proteins are diverse! There are many modification systems that allow 1 gene to code for many proteins. Why do C. elegans & humans have ~ the same number of genes?
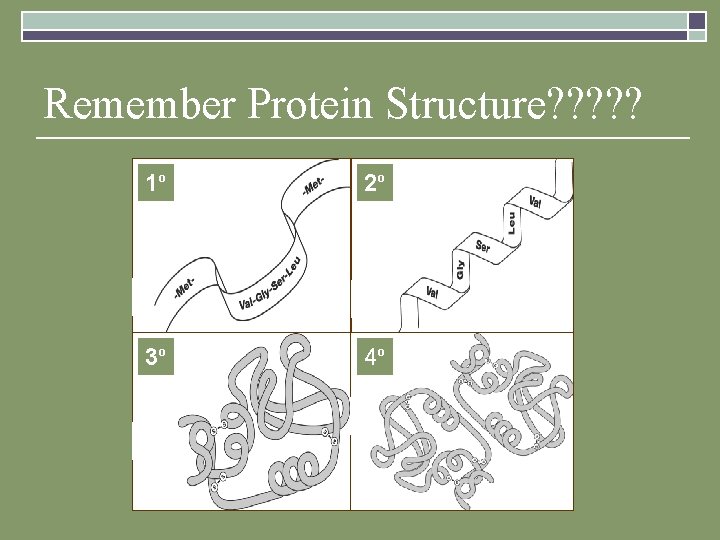
Remember Protein Structure? ? ? 1 o 2 o 3 o 4 o
Posttranscriptional Modifications o o o o RNA Editing Alternative Splicing m. RNA Synthesis & Degradation Proteolytic Cleavage Protein Degradation Protein-Protein Interaction Carbohydrate Modification (Glycosylation) Phosphorylation
RNA Editing o Evolved eukaryotes can change the sequence of m. RNA’s by substituting bases. o This clearly changes the codons and corresponding amino acids. n o Less evolved eukaryotes can delete bases. n n New stop codons New ORF’s New proteins
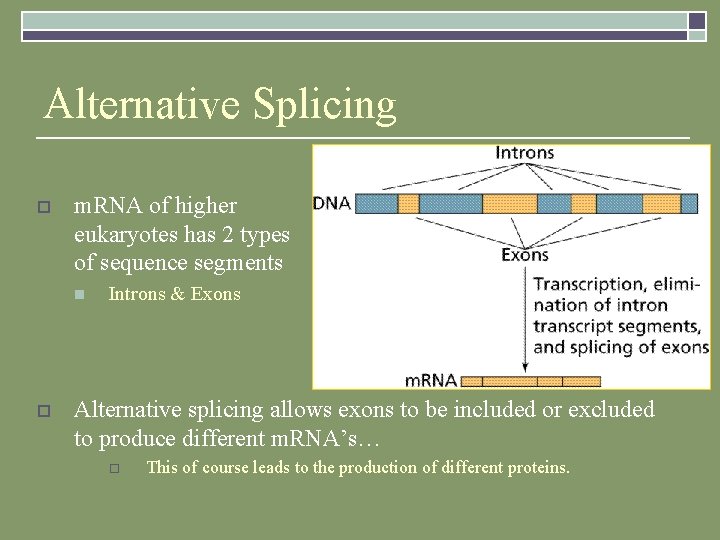
Alternative Splicing o m. RNA of higher eukaryotes has 2 types of sequence segments n o Introns & Exons Alternative splicing allows exons to be included or excluded to produce different m. RNA’s… o This of course leads to the production of different proteins.
m. RNA Synthesis & Degradation o Level of m. RNA synthesis partly determines the level of protein expression o Chemical modifications to m. RNA’s can change their stability and therefore, protein expression levels.

Proteolytic Cleavage o Most proteins undergo cleavage after translation n n o Amino acid encoded by AUG (start codon) o Peptide bonds are broken via protease Usually ‘met’ is removed Misfolded proteins can also undergo proteolytic cleavage
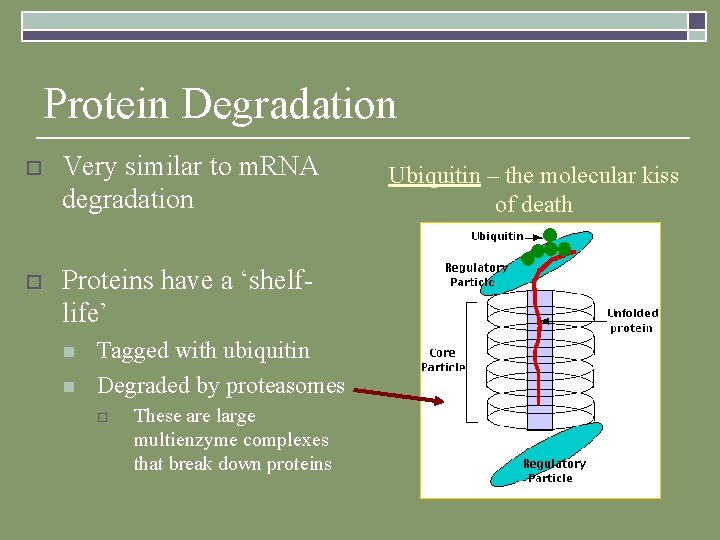
Protein Degradation o Very similar to m. RNA degradation o Proteins have a ‘shelflife’ n n Tagged with ubiquitin Degraded by proteasomes o These are large multienzyme complexes that break down proteins Ubiquitin – the molecular kiss of death
Protein-Protein Interactions o Proteins usually function in complexes. If one protein, necessary for the complex to work properly, is not produced then you may have a non-functional structure.
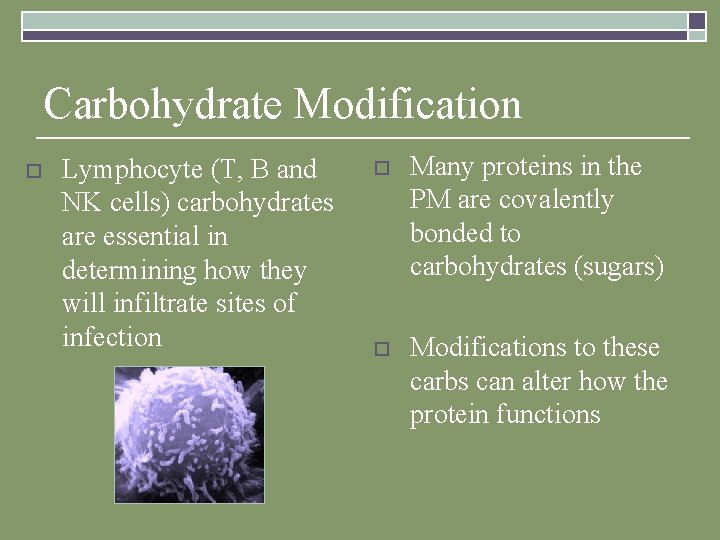
Carbohydrate Modification o Lymphocyte (T, B and NK cells) carbohydrates are essential in determining how they will infiltrate sites of infection o Many proteins in the PM are covalently bonded to carbohydrates (sugars) o Modifications to these carbs can alter how the protein functions
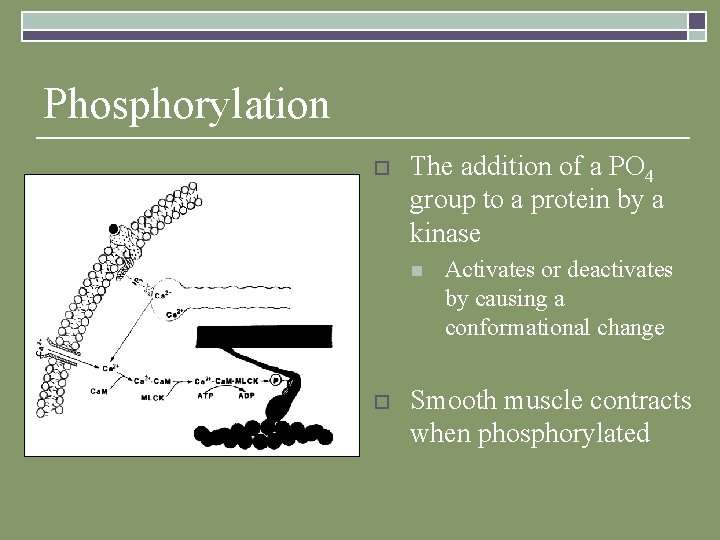
Phosphorylation o The addition of a PO 4 group to a protein by a kinase n o Activates or deactivates by causing a conformational change Smooth muscle contracts when phosphorylated

Hopefully we can see that it becomes increasingly important to examine protein expression & modification among species… That’s what you are doing in lab!


Comparative Proteomics Protein Profiling o. SDS Electrophoresis Physics & Chemistry

Day 2 Day 1 Day 3
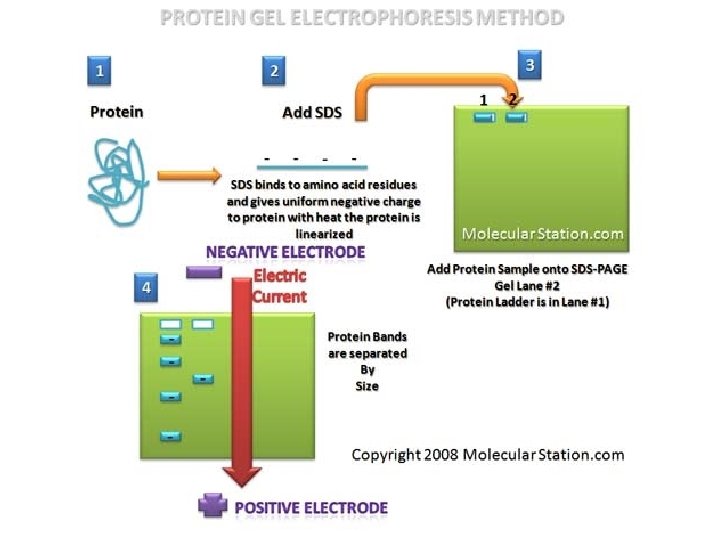
SDS Electrophoresis Physics & Chemistry o SDS-PAGE – sodium dodecyl sulfate polyacrylamide gel electrophoresis n Used to examine proteins o o o How many proteins? Molecular weight? What differences are there in proteins from different sources?

Why SDS-PAGE instead of Agarose? o Gel matrix is polyacrylamide n n o Smaller pores Separates small biomolecules The gel is not uniform in density
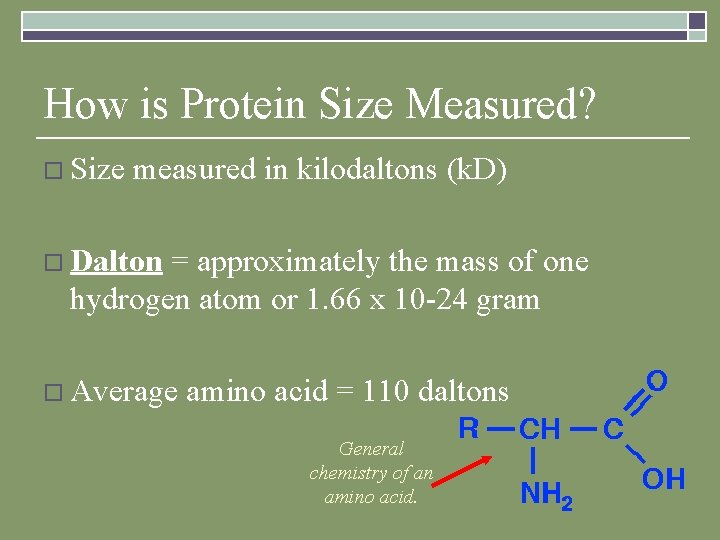
How is Protein Size Measured? o Size measured in kilodaltons (k. D) o Dalton = approximately the mass of one hydrogen atom or 1. 66 x 10 -24 gram o Average amino acid = 110 daltons General chemistry of an amino acid.
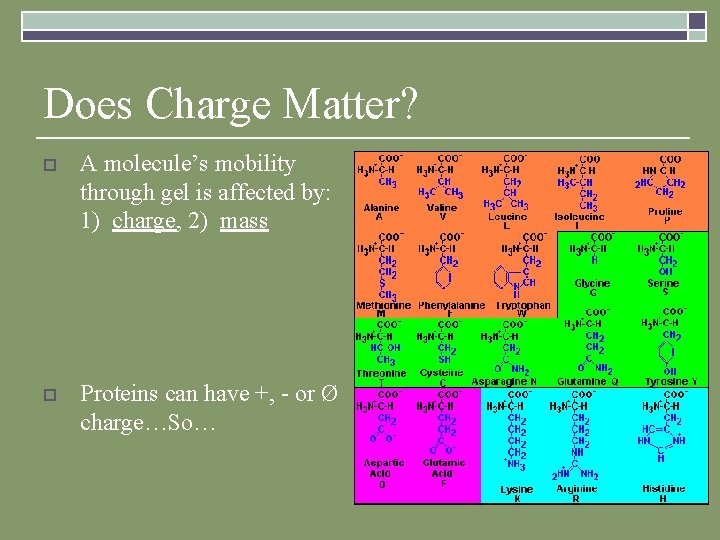
Does Charge Matter? o A molecule’s mobility through gel is affected by: 1) charge, 2) mass o Proteins can have +, - or Ø charge…So…
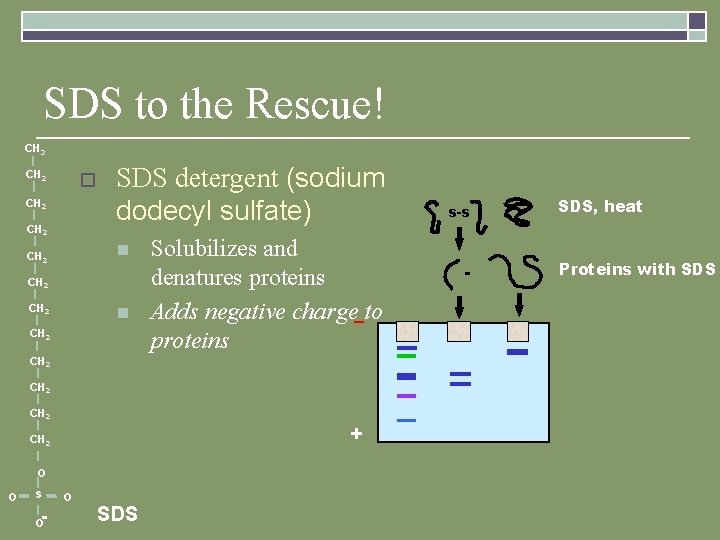
SDS to the Rescue! CH 3 CH 2 o CH 2 SDS detergent (sodium dodecyl sulfate) n CH 2 n CH 2 Solubilizes and denatures proteins Adds negative charge–to proteins CH 2 + CH 2 O - O S SDS s-s SDS, heat Proteins with SDS O O

250 100 75 50 37 25 yo s M n Ac tin & eo rg St u h tfi s Ca t ou Tr on m Sa l rk Sh a P St res an tai da ne rd d s in SDS-Page Example Myosin Heavy Chain Actin Tropomyosin 20 Myosin Light Chains 15 10

Reagents you are working with… 1. 2. 3. 4. 5. Molecular weight marker Laemmli buffer – solubilizes proteins Actin & myosin standard – used as a reference to help ID major conserved muscle proteins & serves as a control DTT – reducing agent that breaks the proteins disulfide bonds Coomassie Stain – will use to stain proteins

Any Questions?
- Slides: 26