Comparative Genomics Overview of the Talk Comparing Genomes








- Slides: 30

Comparative Genomics

Overview of the Talk • Comparing Genomes • Homologies & Families • Sequence Alignments 2

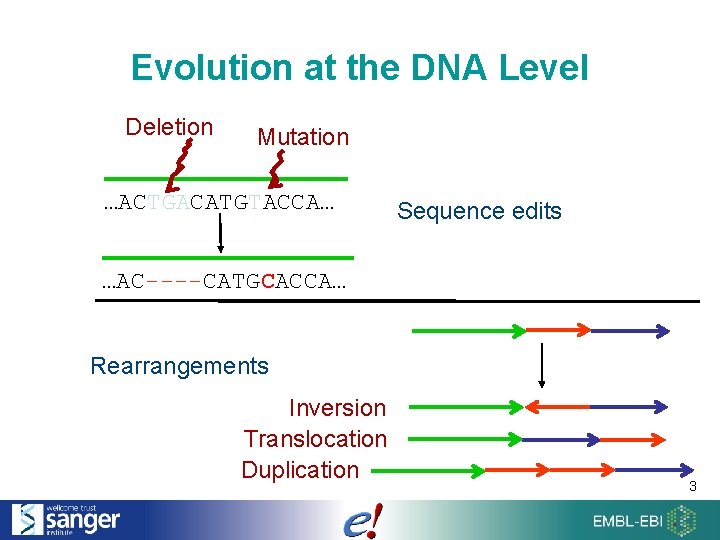
Evolution at the DNA Level Deletion Mutation …ACTGACATGTACCA… Sequence edits …AC----CATGCACCA… Rearrangements Inversion Translocation Duplication 3

Why Compare Genomes? • We can better understand evolution/ speciation • We can find important, functional regions of the sequence (codons, promoters, regulatory regions) • It can help us locate genes in other species that are missing or not well-defined (also through comparison and alignments). 4

Comparing Genomes § Mammals have roughly 3 billion base pairs in their genomes § Over 98% human genes are shared with primates, wth more than 95 -98% similarity between genes. § Even the fruit fly shares 60% of its genes with humans! (March 2000) § Differences: gene structure, sequence Remember… one nucleotide change can cause disease such as sickle cell anemia and cancer. 5

How Does Ensembl Predict Homology? • Uses all the species • Uses a representative protein (the longest) for every gene • Builds a gene tree • Ensembl. Compara Gene. Trees: Analysis of complete, duplication aware phylogenetic trees in vertebrates. Vilella AJ, Severin J, Ureta-Vidal A, Durbin R, Heng L, Birney E. Genome Res. 2008 Nov 24. 6

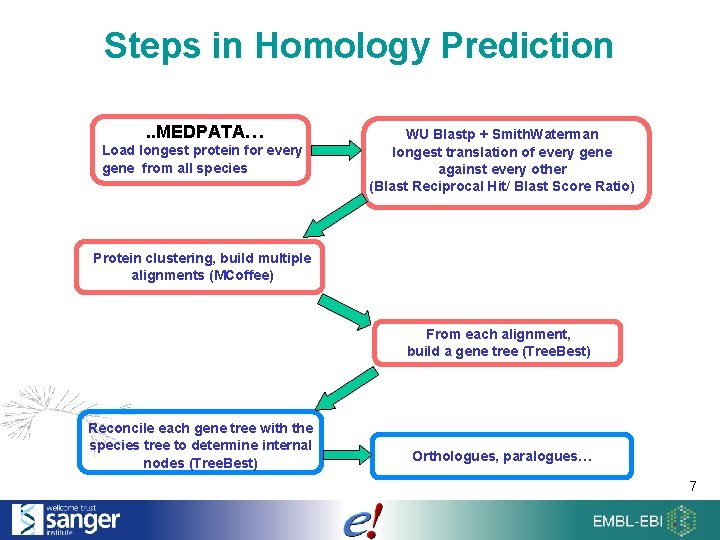
Steps in Homology Prediction. . MEDPATA… Load longest protein for every gene from all species WU Blastp + Smith. Waterman longest translation of every gene against every other (Blast Reciprocal Hit/ Blast Score Ratio) Protein clustering, build multiple alignments (MCoffee) From each alignment, build a gene tree (Tree. Best) Reconcile each gene tree with the species tree to determine internal nodes (Tree. Best) Orthologues, paralogues… 7

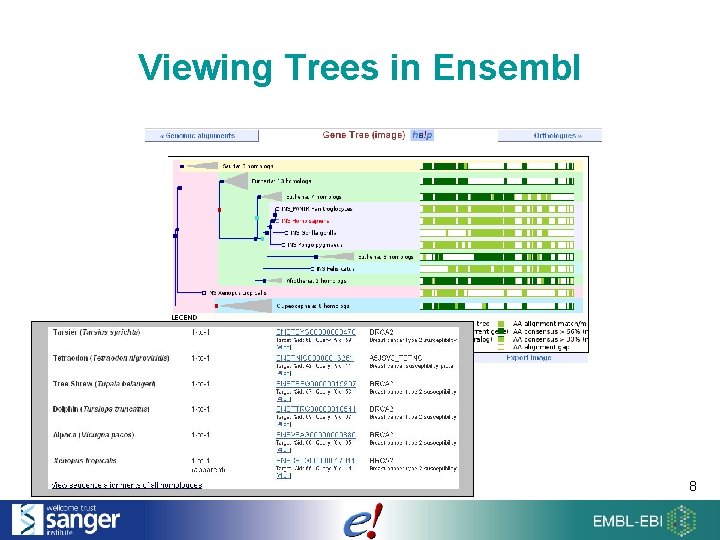
Viewing Trees in Ensembl 8

Types of Homologues • Orthologues : any gene pairwise relation where the ancestor node is a speciation event • Paralogues : any gene pairwise relation where the ancestor node is a duplication event 9

The Gene Tree for INS (insulin precursor) A blue square is a speciation event (Orthologues) A red square is a duplication event (Paralogues) 10

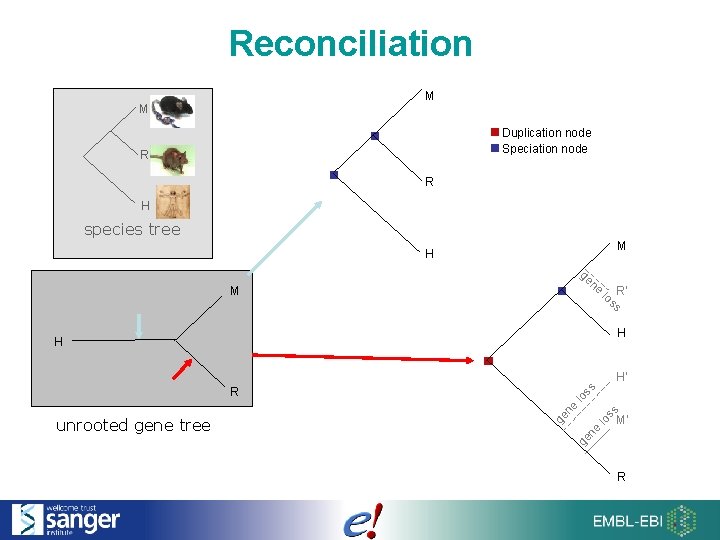
Reconciliation M M Duplication node Speciation node R R H species tree M H ge ne M lo R’ ss H H H’ lo ss ne M’ ge ne lo ge unrooted gene tree ss R R

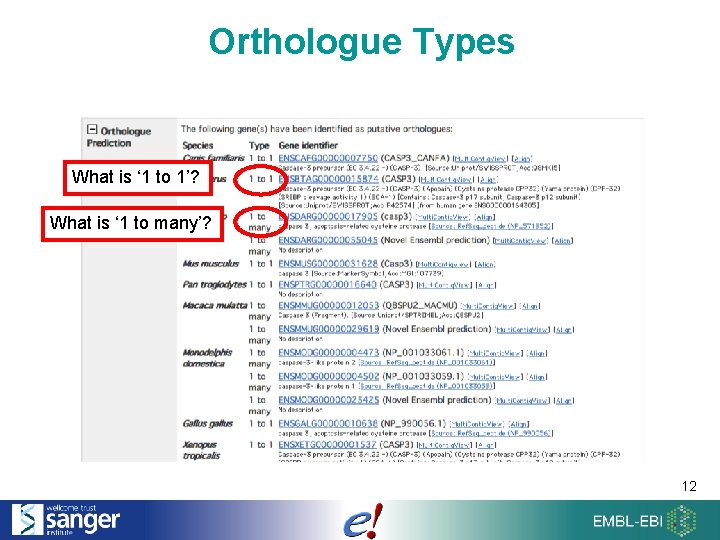
Orthologue Types What is ‘ 1 to 1’? What is ‘ 1 to many’? 12

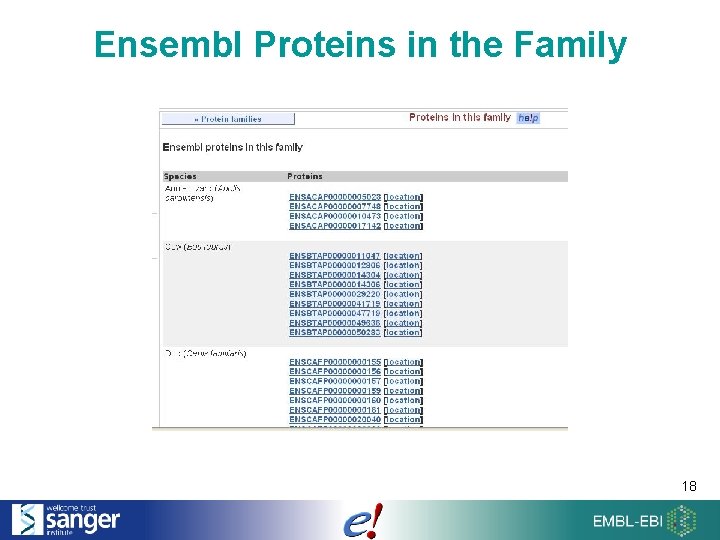
Protein Families • How: Cluster proteins for every isoform in every species + Uni. Prot proteins. • BLASTP comparison of: – all Ensembl ENSP… – all metazoan (animal) proteins in Uni. Prot 13

Homologues Exercise 1. Find the human MYL 6 gene: go to its gene summary. 2. How many paralogues does it have? Find them in the gene tree. 3. Which paralogue is closest to the human MYL 6 gene? In what taxon is the common ancestor? 14

Pan-taxonomic compara Anolis carolinensis Ciona savignyi Danio rerio Equus caballus Gallus gallus Homo sapiens Macaca mulatta Monodelphis domestica Mus musculus Ornithorhynchus anatinus Pan troglodytes Pongo pygmaeus Xenopus tropicalis Anopheles gambiae Caenorhabditis elegans Drosophila melanogaster Dictyostelium discoideum Plasmodium falciparum Plasmodium vivax Arabidopsis thaliana Oryza sativa Vitis vinifera B_aphidicola_Tokyo_1998 B_burgdorferi_DSM_4680 B_subtilis E_coli_K 12 M_tuberculosis_H 37 Rv N_meningitidis_A P_horikoshii S_aureus_N 315 S_pneumoniae_TIGR 4 S_pyogenes_SF 370 W_pipientis_w. Mel Aspergillus nidulans Neurospora crassa Saccharomyces cerevisiae Schizosaccharomyces pombe 15

www. ensemblgenomes. org 16

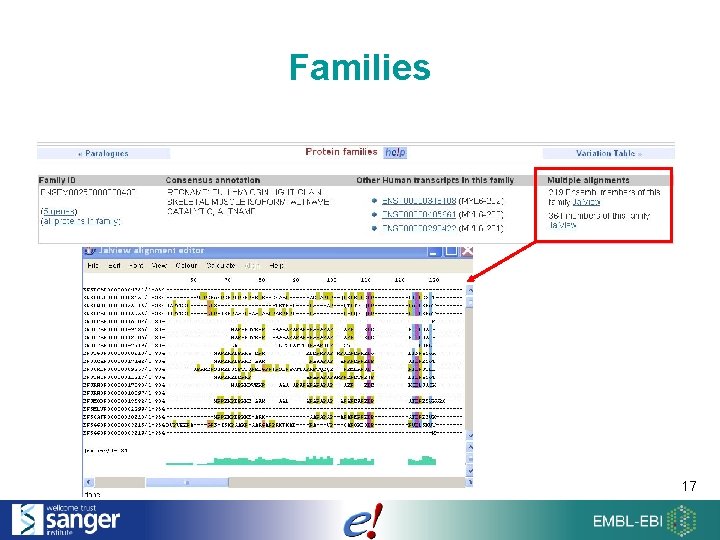
Families 17

Ensembl Proteins in the Family 18

Overview of the Talk • Comparing Genomes • Homologies and Families • Sequence Alignments 19

Aligning Whole Genomes- Why? • To identify homologous regions • To spot trouble gene predictions • Conserved regions could be functional • To define syntenic regions (long regions of DNA sequences where order and orientation is highly conserved) 20

Aligning large genomic sequences Difficulties: • Requires a significant computer resource • Scalability, as more and more genomes are sequenced • Time constraint • As the «true» alignment is not known, then difficult to measure the alignment accuracy and apply the right method 21

Whole Genome Alignments • BLASTZ-net (nucleotide level) closer species e. g. human – mouse • Translated BLAT (amino acid level) more distant species, e. g. human – zebrafish • EPO/PECAN multispecies alignments • ORTHEUS used to determine ancestral alleles 22

Which Multispecies Alignments? Mercator-Pecan • 16 amniota vertebrates + constrained elements Enredo-Pecan-Ortheus (EPO) • For 6 primates • For 5 teleost fish + constrained elements • For 12 eutherian mammals • For 34 eutherian mammals + constrained elements 23

Non-Coding Regions • “Phylogenetic Footprinting” – conserved noncoding regions can be functional • Regulatory regions discovered in this way for genes: Hoxb-1, Hoxb 4, PAX 6, SOX 9 24

More Examples • Highly conserved transcription factor binding sites discovered eg. 401 bp non-coding sequence involved in transcriptional regulation of Interleukins. • New genes (human-mouse comparison) eg. APOA 5, identified as a paralogue to APOA 4 in human and mouse. 25

Going Beyond Mammals Where human-mouse is too conserved, go to other species: Chicken (Mammals and birds: 300 MYA) e. g. A cardiac-specific enhancer of Nkx 2 -5 Human and fish (400 -450 MYA) In 2002, comparison of human to Fugu rubripes led to identification of 1000 genes. 26

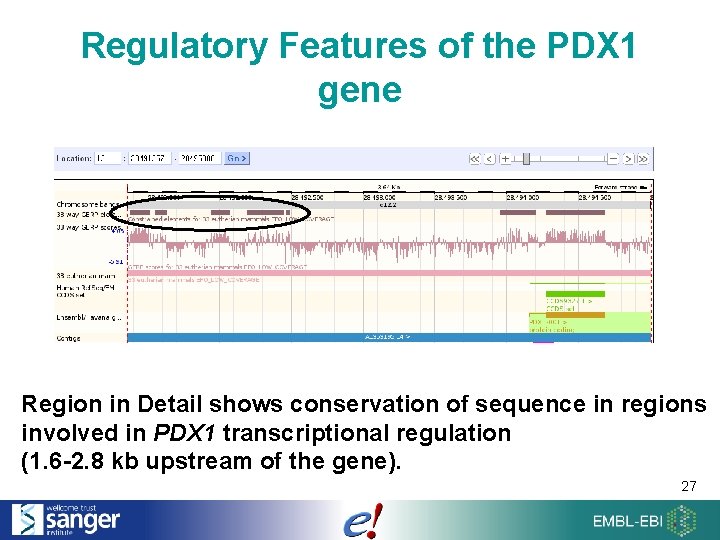
Regulatory Features of the PDX 1 gene Region in Detail shows conservation of sequence in regions involved in PDX 1 transcriptional regulation (1. 6 -2. 8 kb upstream of the gene). 27

Alignments Exercise 1. Have a look at Region in Detail for the ACN 9 gene. 2. Turn on the BLASTZ alignment against macaque. What parts of the macaque genome aligns to this region in human? 3. Turn on the constrained elements for the 33 eutherian mammals. How does this track differ from the BLASTZ alignment? 28

Alignments Continued 1. Zoom out one box in the zoom slide. Are there constrained elements upstream of the ACN 9 transcript that overlap a regulatory feature? 2. View the ‘ 6 primates alignment’ using the Alignments links at the left. 29

Compara Team at EBI • • Javier Herrero Kathryn Beal Stephen Fitzgerald Leo Gordon 30