Comparative genome Hard data and analysis soft interpretations








- Slides: 36

Comparative genome Hard data and analysis soft interpretations ? Peer Bork EMBL & MDC Heidelberg & Berlin bork@embl-heidelberg. de http: //www. bork. embl-heidelberg. de/ www. bork. embl-heidelberg. de

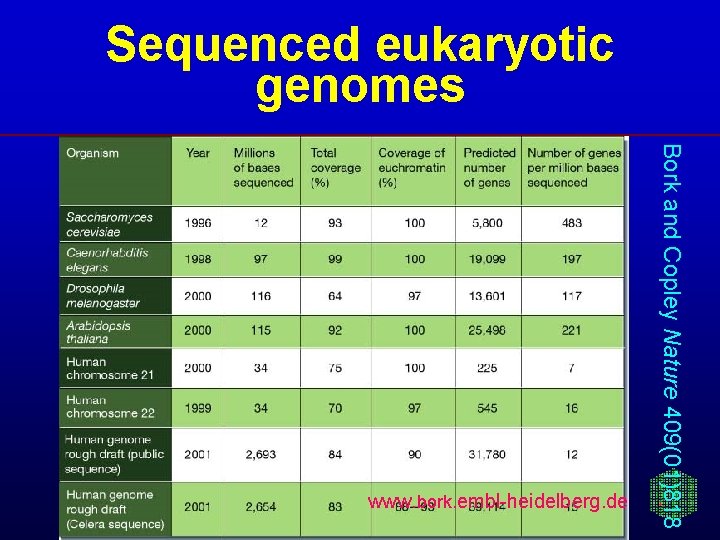
Sequenced eukaryotic genomes Bork and Copley Nature 409(01)818 www. bork. embl-heidelberg. de

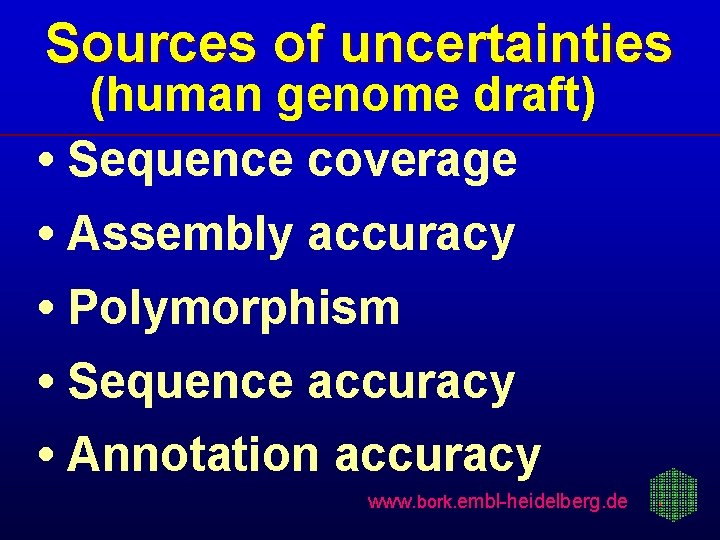
Sources of uncertainties (human genome draft) • Sequence coverage • Assembly accuracy • Polymorphism • Sequence accuracy • Annotation accuracy www. bork. embl-heidelberg. de

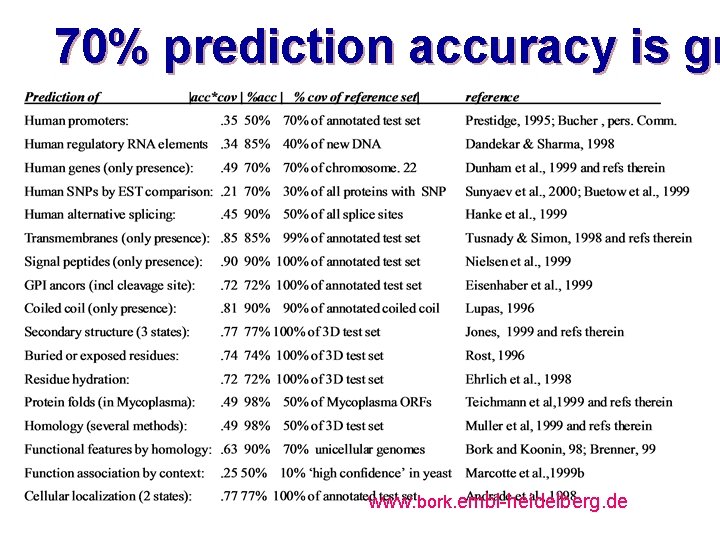
70% prediction accuracy is gr www. bork. embl-heidelberg. de

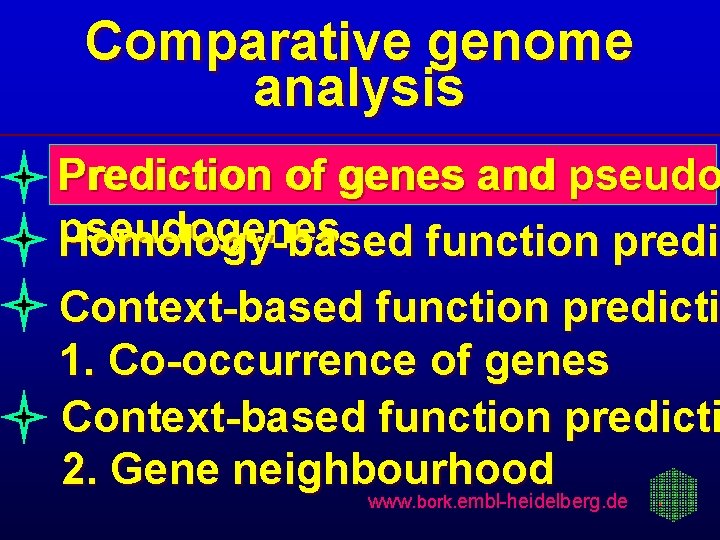
Comparative genome analysis Prediction of genes and pseudogenes Homology-based function predic predi Context-based function predicti 1. Co-occurrence of genes Context-based function predicti 2. Gene neighbourhood www. bork. embl-heidelberg. de

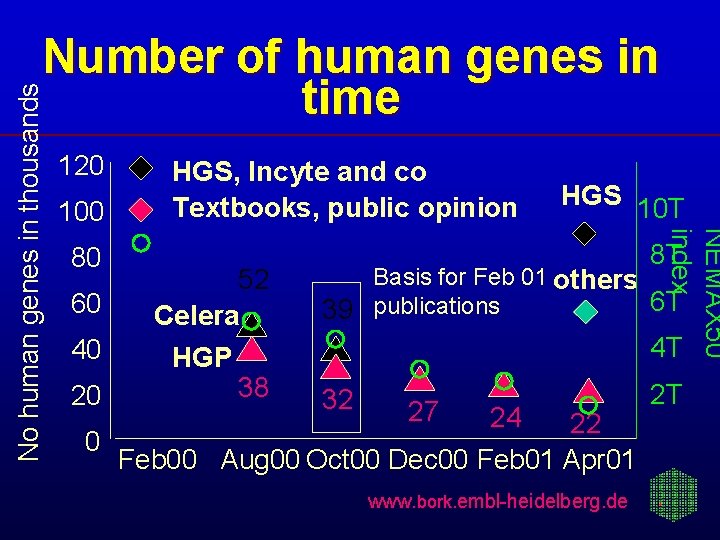
120 100 80 60 40 20 HGS, Incyte and co Textbooks, public opinion 52 Celera HGP 38 HGS 10 T NEMAX 50 index No human genes in thousands Number of human genes in time 8 T Basis for Feb 01 others 6 T 39 publications 4 T 32 27 24 22 0 Feb 00 Aug 00 Oct 00 Dec 00 Feb 01 Apr 01 www. bork. embl-heidelberg. de 2 T

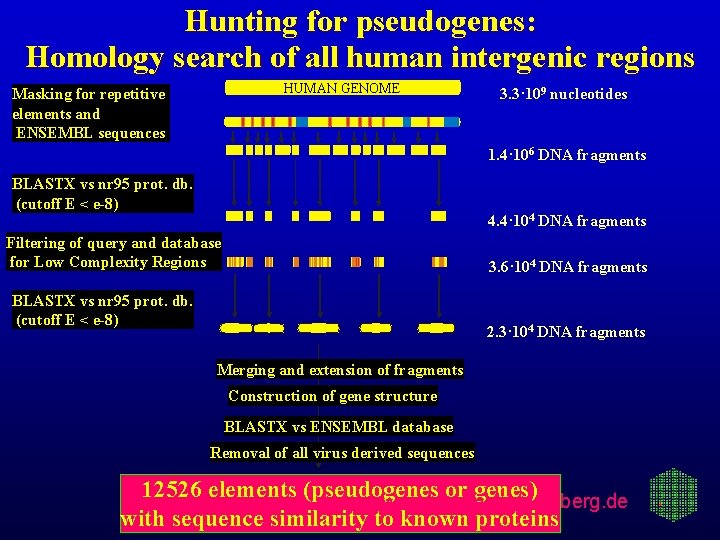
Hunting for pseudogenes: Homology search of all human intergenic regions HUMAN GENOME Masking for repetitive elements and ENSEMBL sequences 3. 3· 109 nucleotides 1. 4· 106 DNA fragments BLASTX vs nr 95 prot. db. (cutoff E < e-8) 4. 4· 104 DNA fragments Filtering of query and database for Low Complexity Regions 3. 6· 104 DNA fragments BLASTX vs nr 95 prot. db. (cutoff E < e-8) 2. 3· 104 DNA fragments Merging and extension of fragments Construction of gene structure BLASTX vs ENSEMBL database Removal of all virus derived sequences 12526 elements (pseudogenes or genes) www. bork. embl-heidelberg. de with sequence similarity to known proteins

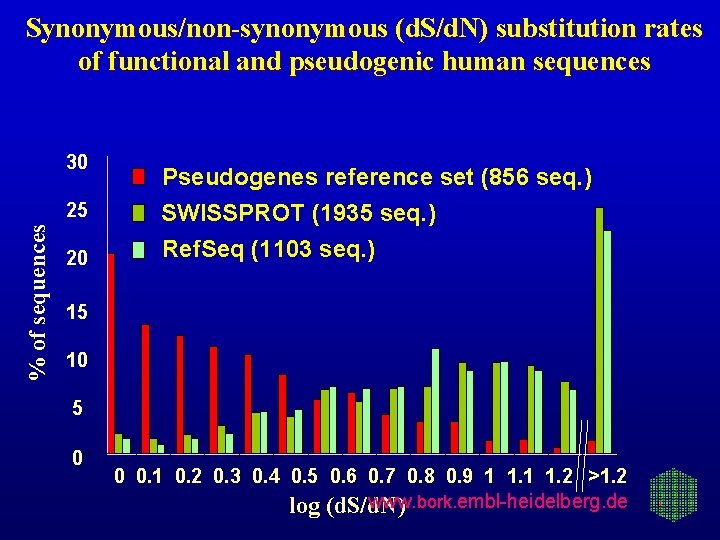
Synonymous/non-synonymous (d. S/d. N) substitution rates of functional and pseudogenic human sequences 30 % of sequences 25 20 Pseudogenes reference set (856 seq. ) SWISSPROT (1935 seq. ) Ref. Seq (1103 seq. ) 15 10 5 0 0 0. 1 0. 2 0. 3 0. 4 0. 5 0. 6 0. 7 0. 8 0. 9 1 1. 2 >1. 2 www. bork. embl-heidelberg. de log (d. S/d. N)

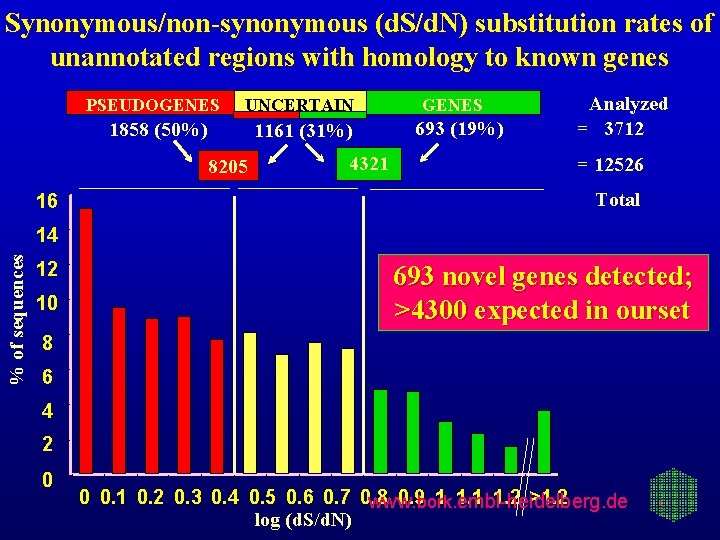
Synonymous/non-synonymous (d. S/d. N) substitution rates of unannotated regions with homology to known genes PSEUDOGENES UNCERTAIN 1858 (50%) 1161 (31%) 8205 16 4321 GENES 693 (19%) Analyzed = 3712 = 12526 Total % of sequences 14 12 10 693 novel genes detected; >4300 expected in ourset 8 6 4 2 0 0 0. 1 0. 2 0. 3 0. 4 0. 5 0. 6 0. 7 0. 8 0. 9 bork 1. embl-heidelberg. de 1. 1 1. 2 >1. 2 www. log (d. S/d. N)

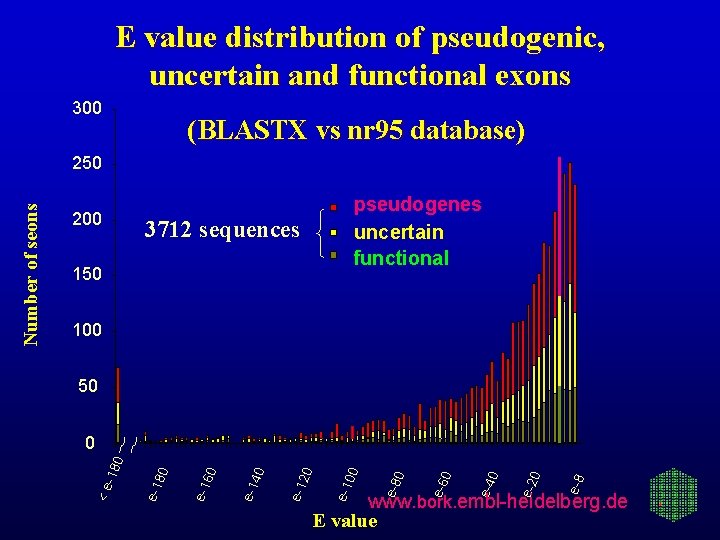
E value distribution of pseudogenic, uncertain and functional exons 300 (BLASTX vs nr 95 database) 200 pseudogenes uncertain functional 3712 sequences 150 100 50 e-8 0 e-2 0 e-4 0 e-6 0 e-8 00 e-1 20 e-1 40 e-1 60 e-1 80 e-1 -18 0 0 <e Number of seons 250 www. bork. embl-heidelberg. de E value

Comparative genome analysis Prediction of genes and pseudogenes Homology-based function predic predi Context-based function predicti 1. Co-occurrence of genes Context-based function predicti 2. Gene neighbourhood www. bork. embl-heidelberg. de

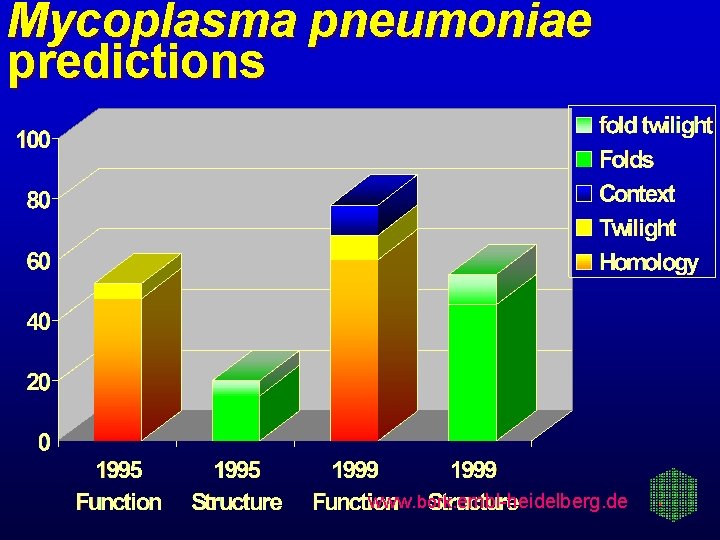
Mycoplasma pneumoniae predictions www. bork. embl-heidelberg. de

Molecular Functions have to be defined on a domain basis i. e. separately for each structurally independent unit within Henikoff etaal. 1997 www. bork. embl-heidelberg. de Science 278, 609 sequence

www. bork. embl-heidelberg. de

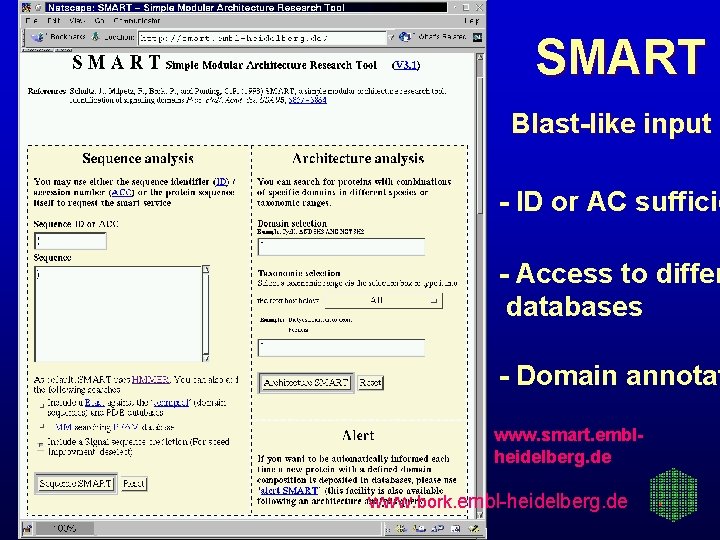
SMART Blast-like input - ID or AC sufficie - Access to differ databases - Domain annotat www. smart. emblheidelberg. de www. bork. embl-heidelberg. de

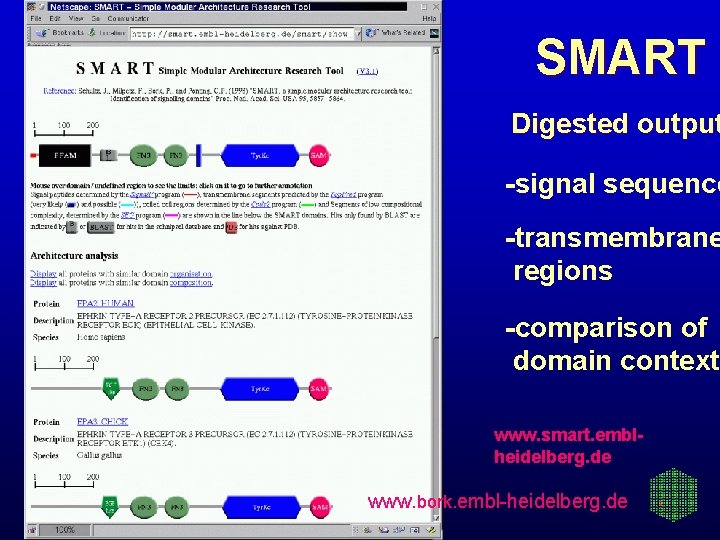
SMART Digested output -signal sequence -transmembrane regions -comparison of domain context www. smart. emblheidelberg. de www. bork. embl-heidelberg. de

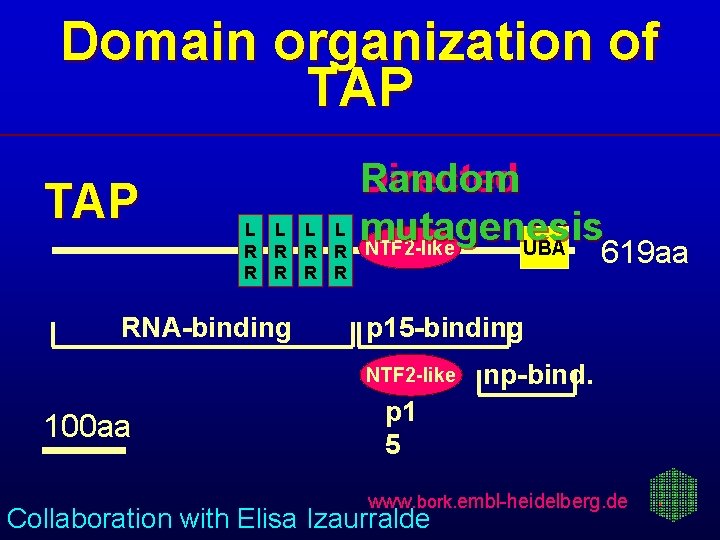
Domain organization of TAP L L R R R R RNA-binding Random Directed mutagenesis NTF 2 -like UBA 619 aa p 15 -binding NTF 2 -like 100 aa np-bind. p 1 5 www. bork. embl-heidelberg. de Collaboration with Elisa Izaurralde

Directed mutagenesis confirms predicted TAP/p 15 interaction Red - loss of binding Blue - no effect on binding Gray - alanine sca

Top 10 domains* in human Species man fly wormyeast cress Total no genes 13300 18200 6100 25700 26500(26500) 1 64 0 Immunoglobulin 765(381) 140 115 48 C 2 H 2 zinc finger 706(607) 357 151 Protein kinase 575(501) 319 437 121 1049 16 0 Rhod. -like GPCR 569(616) 97 358 331 198 183 97 P-loop NTPase 433 80 10 50 6 Rev. transcriptase 350 255 96 54 300(224) 157 RRM (RNA-binding) 210 91 WD 40 (G-protein)277(136) 162 102 19 120 Ankyrin repeat 276(145) 105 107 109 9 118 267(160) 148 Homeobox www. bork. embl-heidelberg. de *Only no of genes given, no of domains higher; note that only around Nature 409 (01)860; Science 291(01)1304 90% is sequenced

Comparative genome analysis Prediction of genes and pseudo Homology-based function predic predi Context-based function predicti 1. Co-occurrence of genes Context-based function predicti 2. Gene neighbourhood www. bork. embl-heidelberg. de

Function prediction via genomic context information Gene context: - Gene fusion as distinct neighborhood subset - Conserved gene neighborhood in genomes - Conserved co-occurrence of genes in species (‘phylogentic profile’, ‘COG pattern’) - Surrounding and shared regulatory elements Knowledge-based context: - Pathway data (can overrule homology!) - Gene expression data (co-expression etc. ) - Protein interaction /localisation www. bork. embl-heidelberg. de - Scientific literature

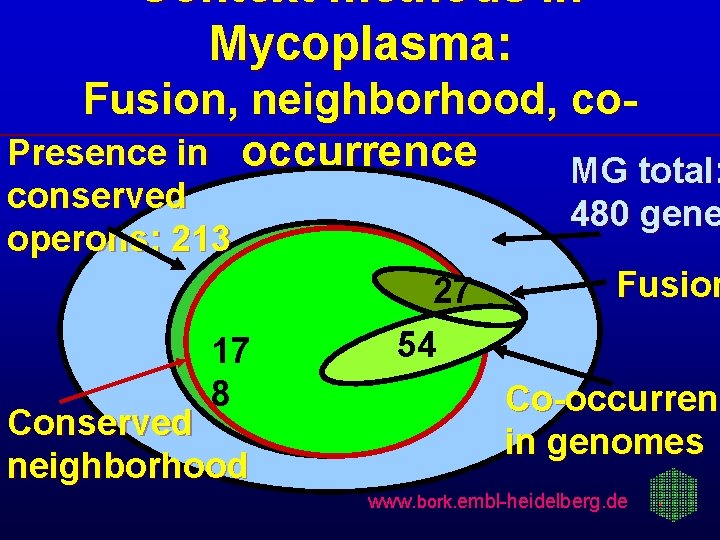
Context methods in Mycoplasma: Fusion, neighborhood, co. Presence in occurrence MG total: conserved operons: 213 17 8 Conserved neighborhood 480 gene 27 54 Fusion Co-occurrenc Co-occurren in genomes www. bork. embl-heidelberg. de

Orthology vs paralogy … within homology paralogy gene A 1 gene A 2 Genome A gene B 2 Genome B orthology gene B 1 history gene 1 gene 2 gene A 1 gene B 1 gene A 2 gene B 2 www. bork. embl-heidelberg. de

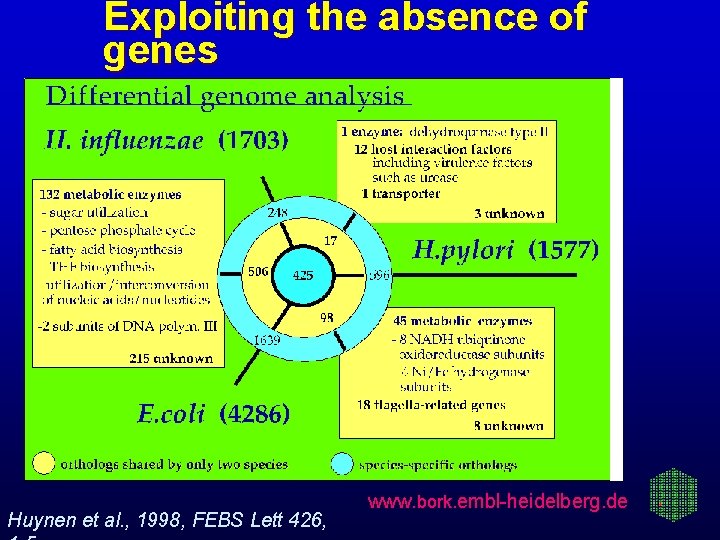
Exploiting the absence of genes Huynen et al. , 1998, FEBS Lett 426, www. bork. embl-heidelberg. de

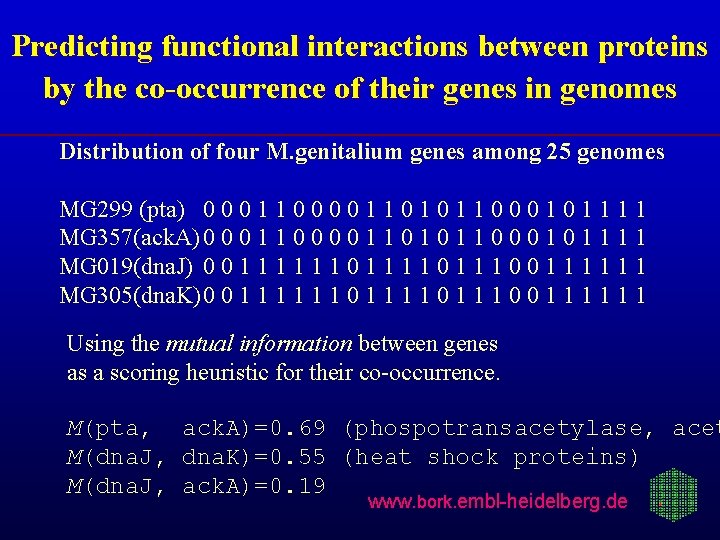
Predicting functional interactions between proteins by the co-occurrence of their genes in genomes Distribution of four M. genitalium genes among 25 genomes MG 299 (pta) 0 0 0 1 1 0 1 0 1 1 0 0 0 1 1 1 1 MG 357(ack. A) 0 0 0 1 1 0 1 0 1 1 0 0 0 1 1 1 1 MG 019(dna. J) 0 0 1 1 1 0 1 1 1 0 0 1 1 1 MG 305(dna. K)0 0 1 1 1 0 1 1 1 0 0 1 1 1 Using the mutual information between genes as a scoring heuristic for their co-occurrence. M(pta, ack. A)=0. 69 (phospotransacetylase, acet M(dna. J, dna. K)=0. 55 (heat shock proteins) M(dna. J, ack. A)=0. 19 www. bork. embl-heidelberg. de

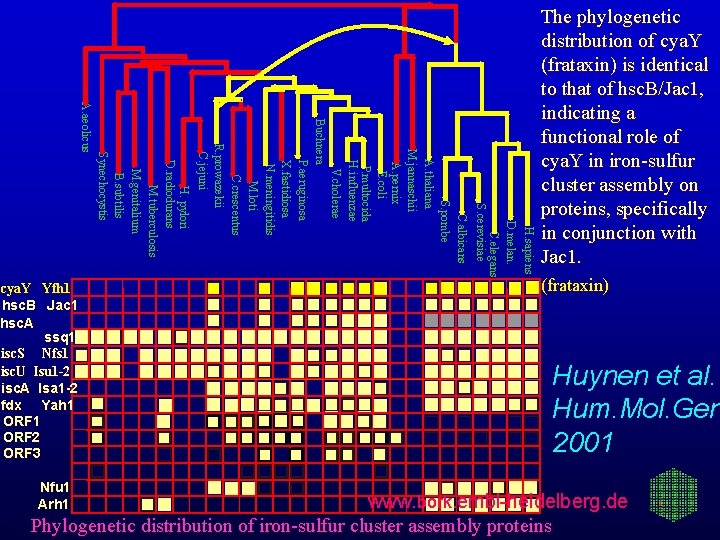
H. sapiens D. melan. C. elegans S. cerevisiae Nfu 1 Arh 1 C. albicans S. pombe A. thaliana M. jannaschii A. pernix E. coli P. multocida H. influenzae V. cholerae Buchnera P. aeruginosa X. fastidiosa N. meningitidis M. loti C. crescentus R. prowazekii C. jejuni H. pylori D. radiodurans M. tuberculosis M. genitalium B. subtilis Synechocystis A. aeolicus cya. Y Yfh 1 hsc. B Jac 1 hsc. A ssq 1 isc. S Nfs 1 isc. U Isu 1 -2 isc. A Isa 1 -2 fdx Yah 1 ORF 2 ORF 3 The phylogenetic distribution of cya. Y (frataxin) is identical to that of hsc. B/Jac 1, indicating a functional role of cya. Y in iron-sulfur cluster assembly on proteins, specifically in conjunction with Jac 1. (frataxin) Huynen et al. Hum. Mol. Gen 2001 www. bork. embl-heidelberg. de Phylogenetic distribution of iron-sulfur cluster assembly proteins

Comparative genome analysis Prediction of genes and pseudo Homology-based function predic predi Context-based function predicti 1. Co-occurrence of genes Context-based function predicti 2. Gene neighbourhood www. bork. embl-heidelberg. de

Genome alignment www. bork. embl-heidelberg. de

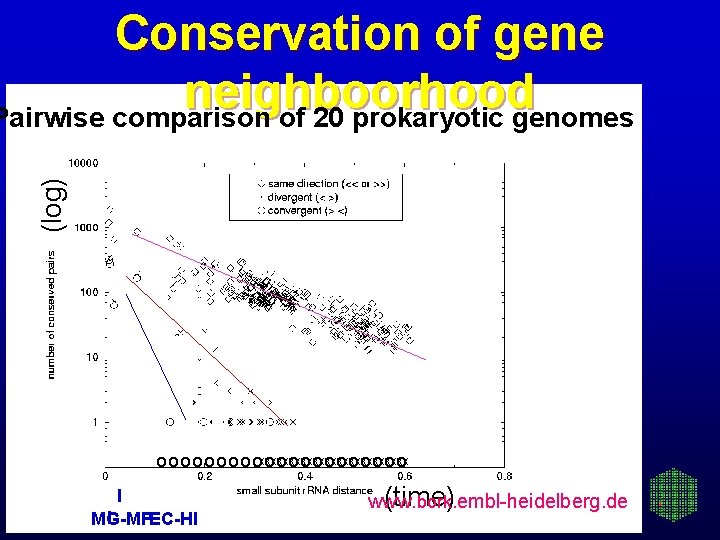
(log) Conservation of gene neighboorhood Pairwise comparison of 20 prokaryotic genomes o o o o oxxxxxxxxxxxxx oooooo I I MG-MP EC-HI (time) www. bork. embl-heidelberg. de

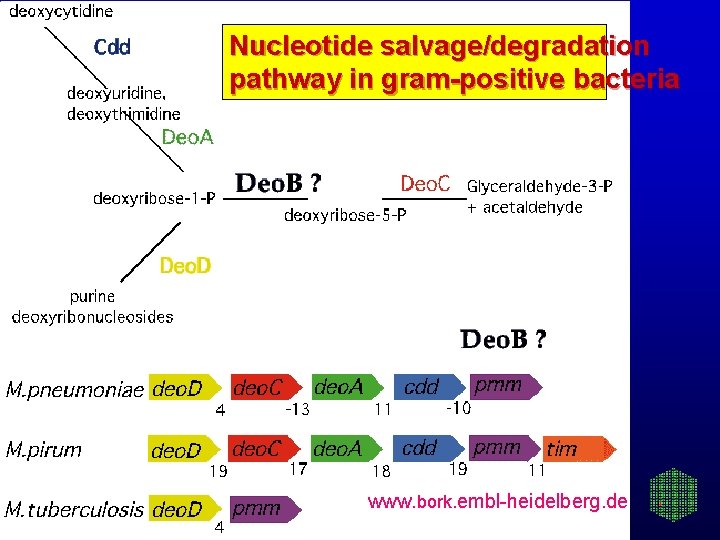
Nucleotide salvage/degradation pathway in gram-positive bacteria www. bork. embl-heidelberg. de

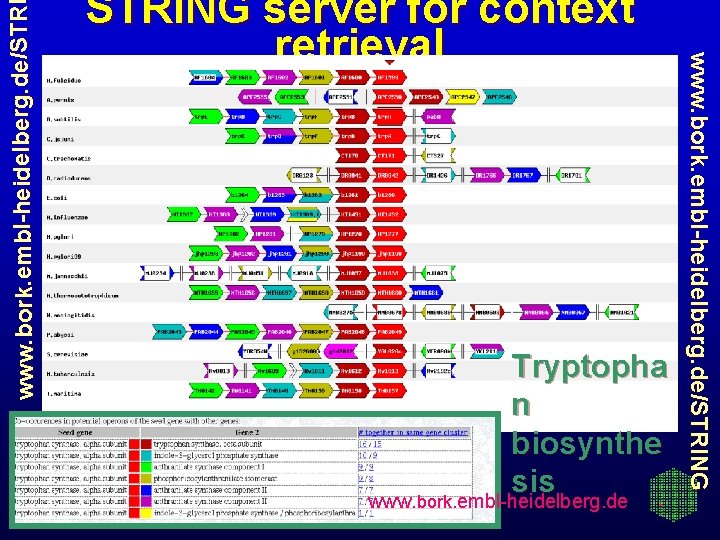
www. bork. embl-heidelberg. de/STR Tryptopha n biosynthe sis www. bork. embl-heidelberg. de/STRING server for context retrieval

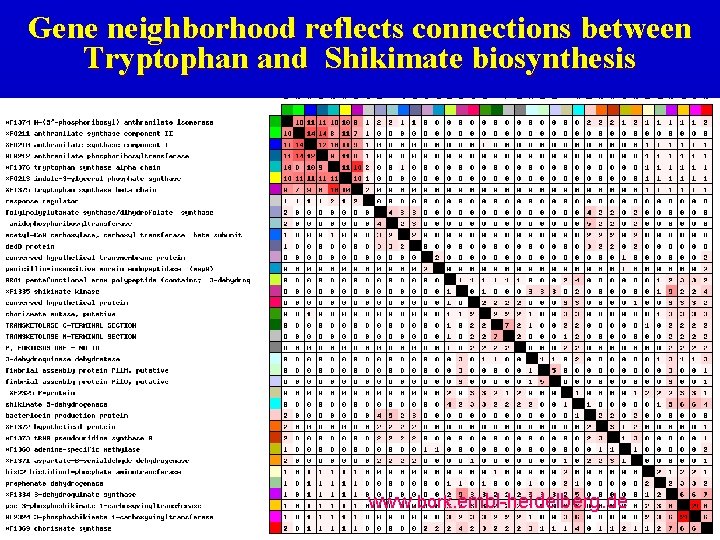
Gene neighborhood reflects connections between Tryptophan and Shikimate biosynthesis www. bork. embl-heidelberg. de

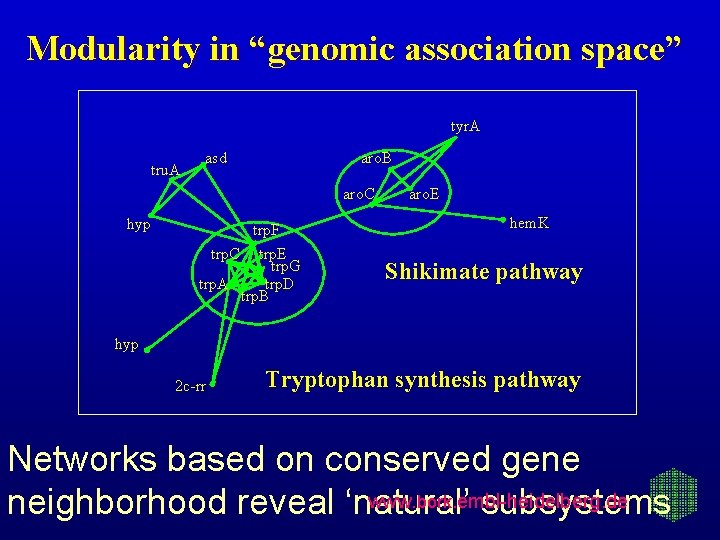
Modularity in “genomic association space” tyr. A tru. A asd aro. B aro. C hyp trp. F trp. C trp. E trp. G trp. A trp. D trp. B aro. E hem. K Shikimate pathway hyp 2 c-rr Tryptophan synthesis pathway Networks based on conserved gene www. bork. embl-heidelberg. de neighborhood reveal ‘natural’ subsystems

(pseudo)genes Yan Yuan Mikita Suyama David Torrents www. bork. embl-heidelberg. de

SMART Ivica Letuni Rich Copley www. bork. embl-heidelberg. de

www. bork. embl-heidelberg. d *Frank (D) Yan (C) Peer (D) *Martijn (NL) *Gert (D) Shamil (RU) *Vassily (RU) *Birgit (D) Tobias (D) Richard (UK) *Luis (E) Mikita (J) Miguel (E) *Jörg (D) *left EMB Warren (US) Berend (NL) David (E), Ivica (Hr), Caroline (E), Steffen(D), www. bork. embl-heidelberg. de Francesca(I), Jan (D), Parantu(In), Christian(D)