Community Detection in Graphs Finding overlaps Facebook Network

Community Detection in Graphs: Finding overlaps
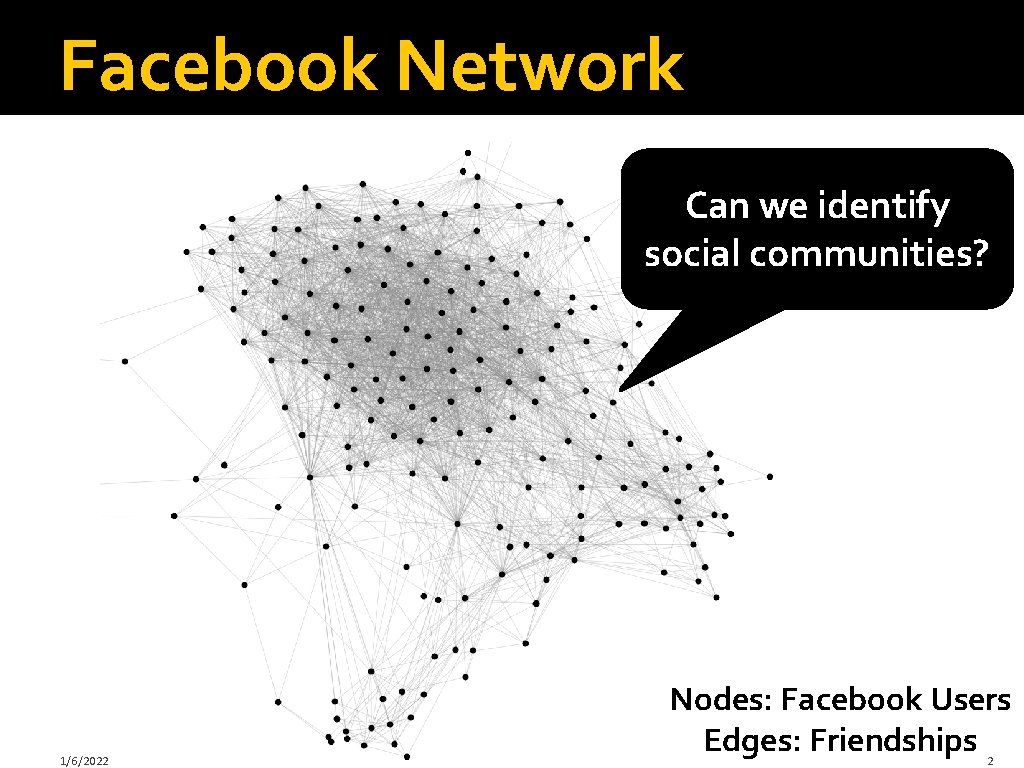
Facebook Network Can we identify social communities? 1/6/2022 Nodes: Facebook Users Edges: Friendships 2
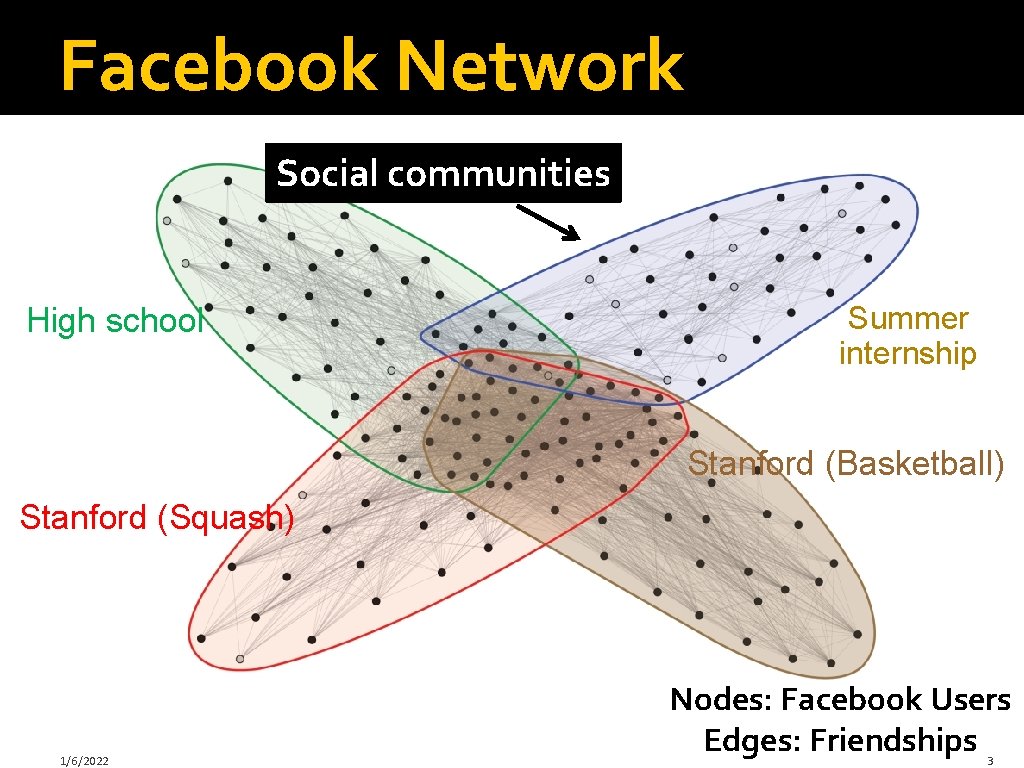
Facebook Network Social communities High school Summer internship Stanford (Basketball) Stanford (Squash) 1/6/2022 Nodes: Facebook Users Edges: Friendships 3
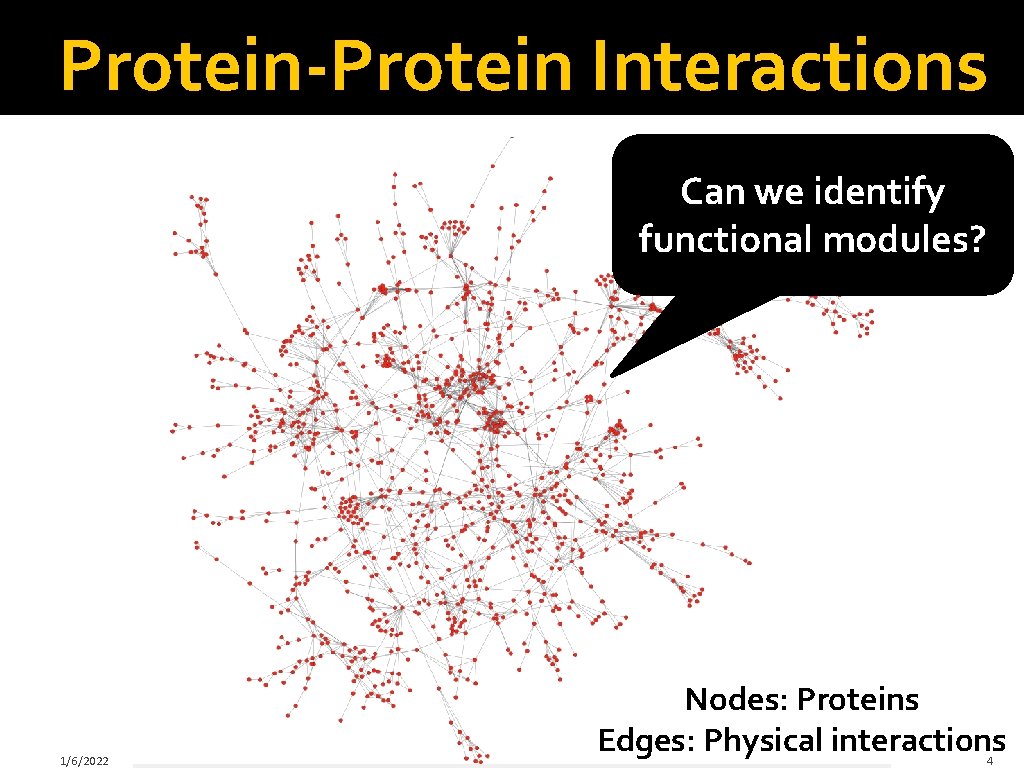
Protein-Protein Interactions Can we identify functional modules? 1/6/2022 Nodes: Proteins Edges: Physical interactions 4
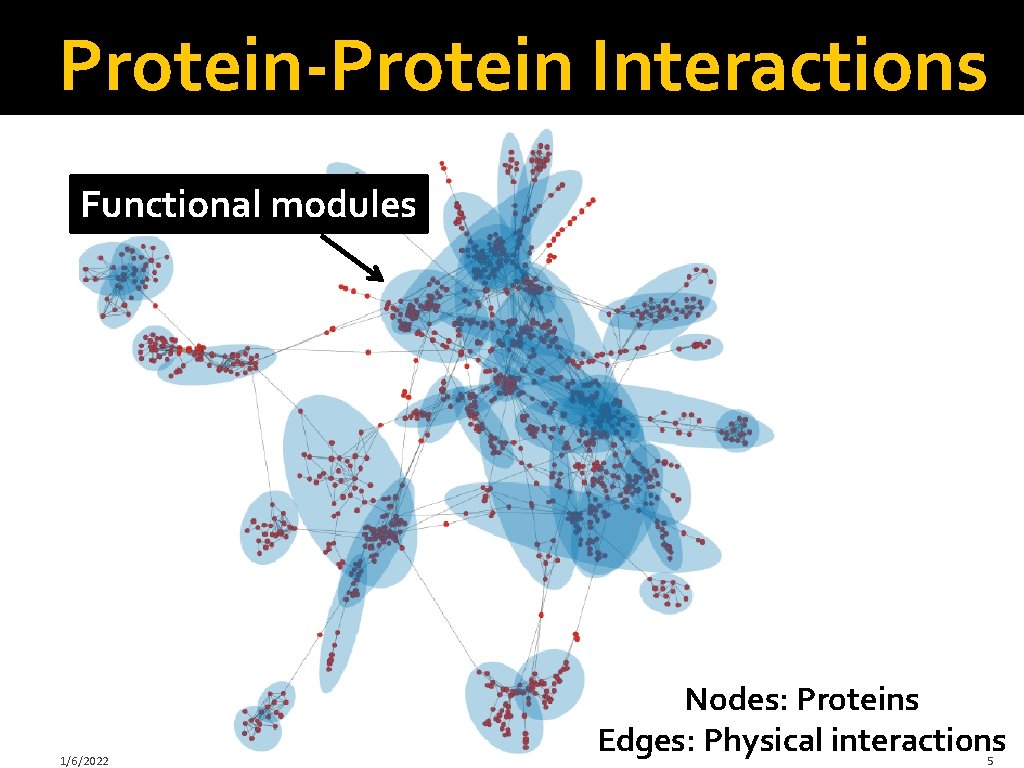
Protein-Protein Interactions Functional modules 1/6/2022 Nodes: Proteins Edges: Physical interactions 5
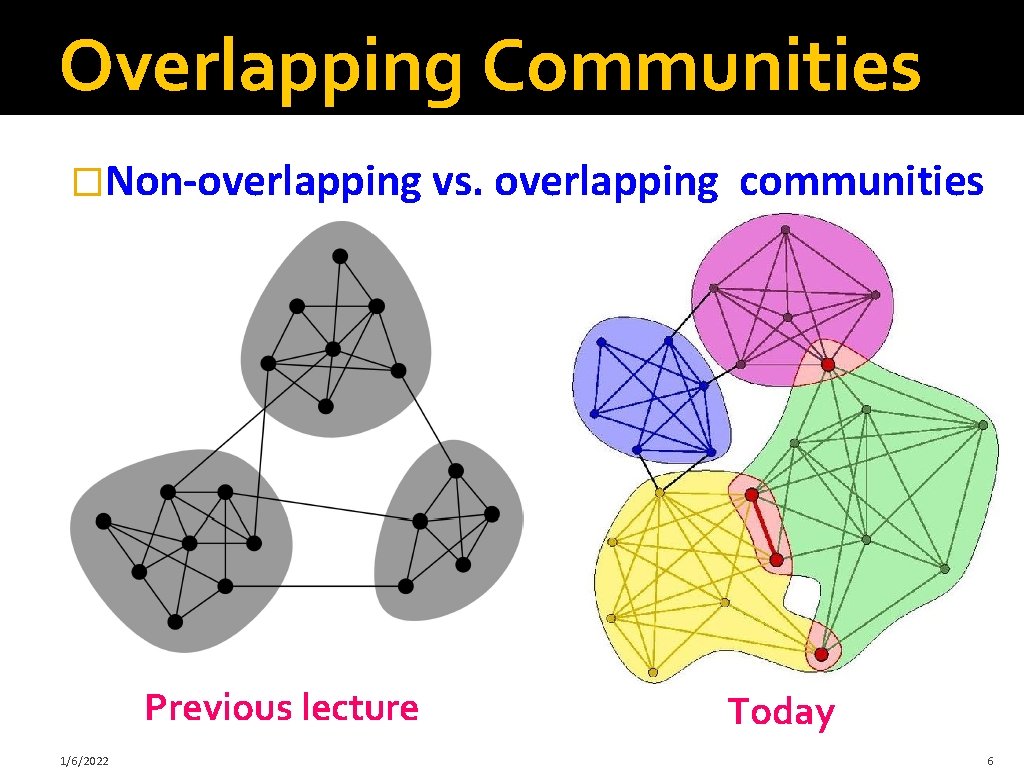
Overlapping Communities �Non-overlapping vs. overlapping Previous lecture 1/6/2022 communities Today 6
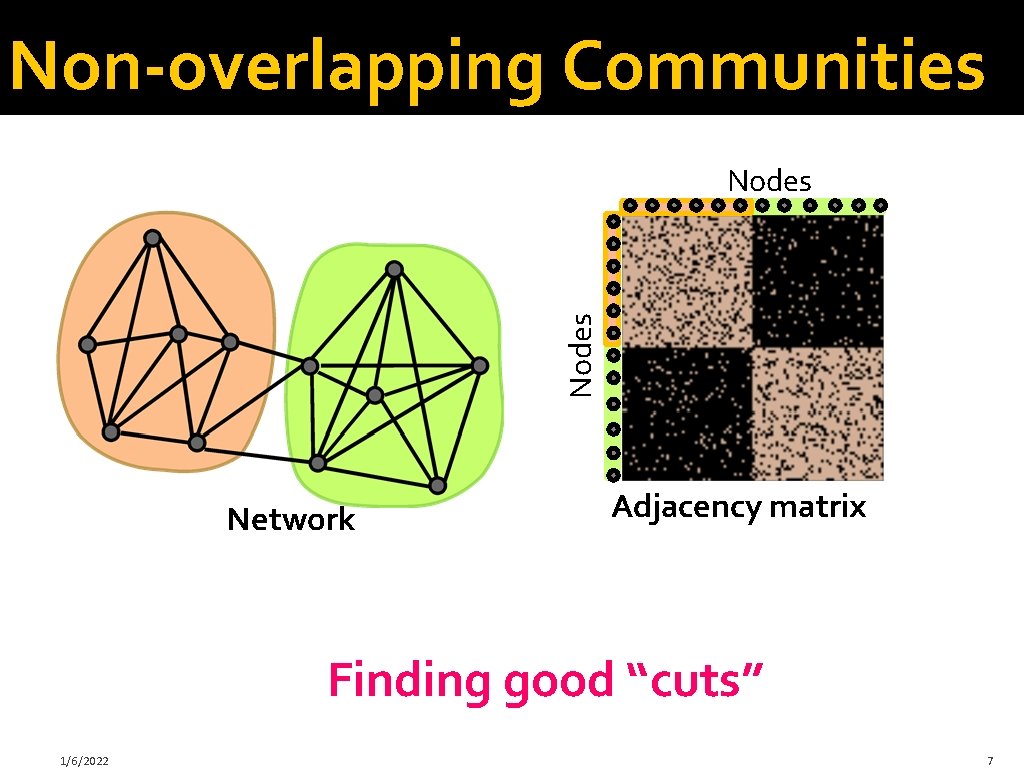
Non-overlapping Communities Nodes Network Adjacency matrix Finding good “cuts” 1/6/2022 7

Communities as Tiles! What if communities overlap? Communities as “tiles” 1/6/2022 8
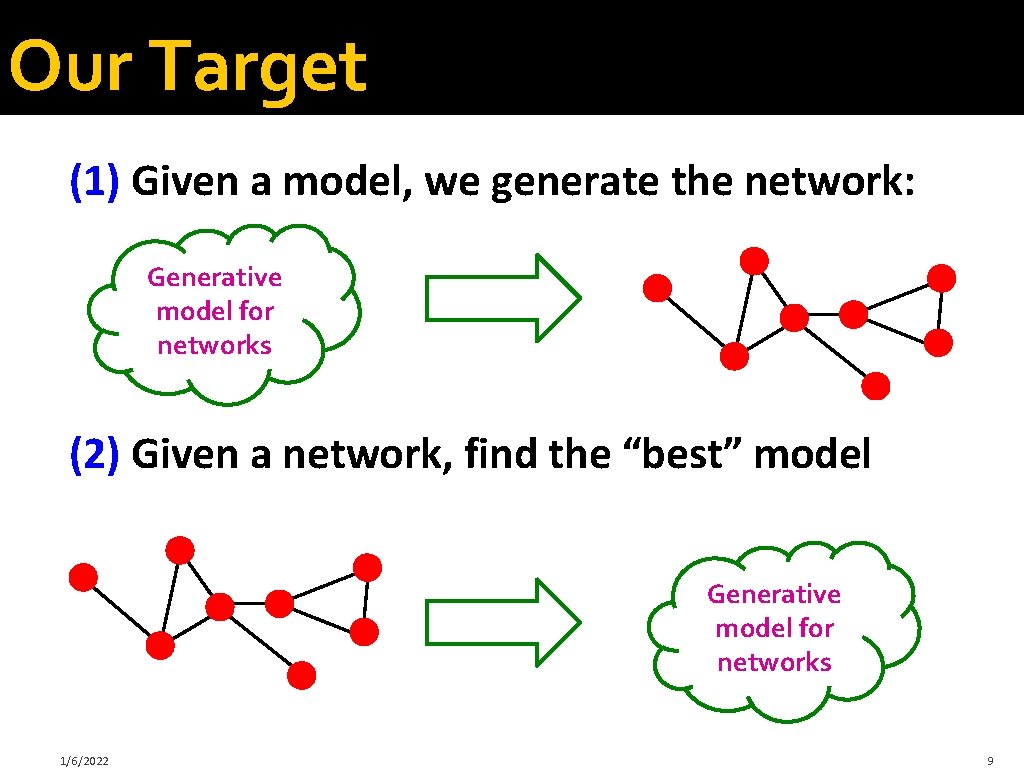
Our Target (1) Given a model, we generate the network: B Generative model for networks F A D E G C H (2) Given a network, find the “best” model B F A D E G C H 1/6/2022 Generative model for networks 9

Model of networks �Goal: Define a model for generating networks § The model will have a set of “parameters” that we will later want to estimate (to detect communities) Generative model for networks B F A D E G C H �Q: Given a set of nodes and their community memberships, how do communities “generate” edges of the network? 1/6/2022 10
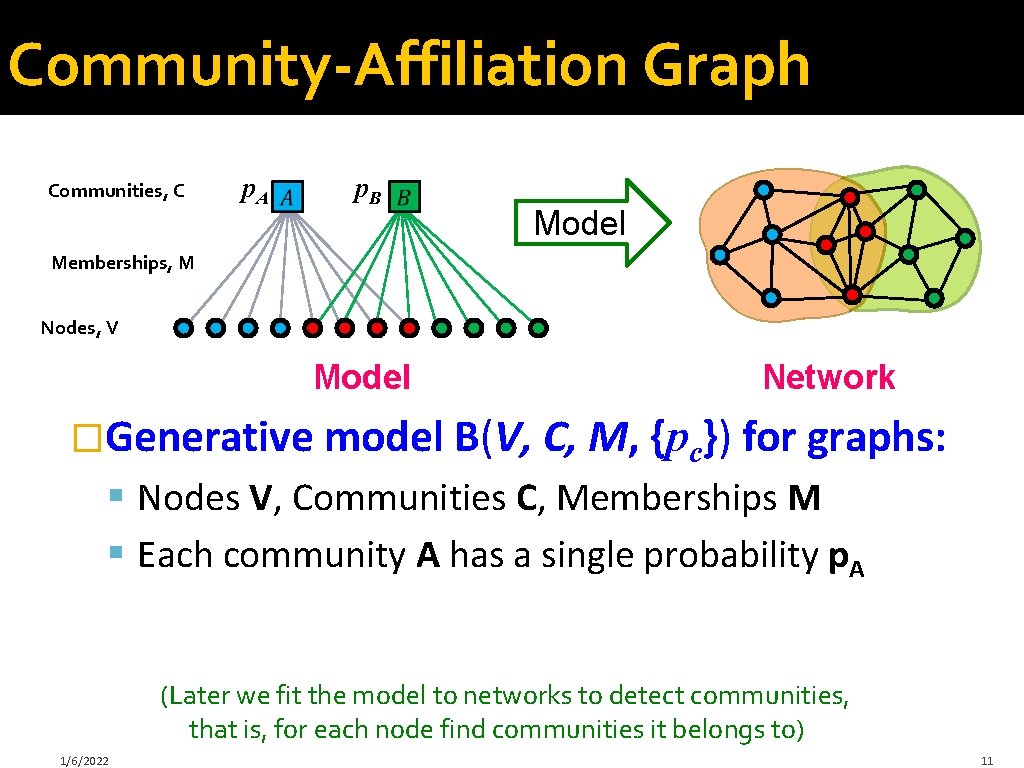
Community-Affiliation Graph Communities, C p. A p. B Model Memberships, M Nodes, V Model Network �Generative model B(V, C, M, {pc}) for graphs: § Nodes V, Communities C, Memberships M § Each community A has a single probability p. A (Later we fit the model to networks to detect communities, that is, for each node find communities it belongs to) 1/6/2022 11
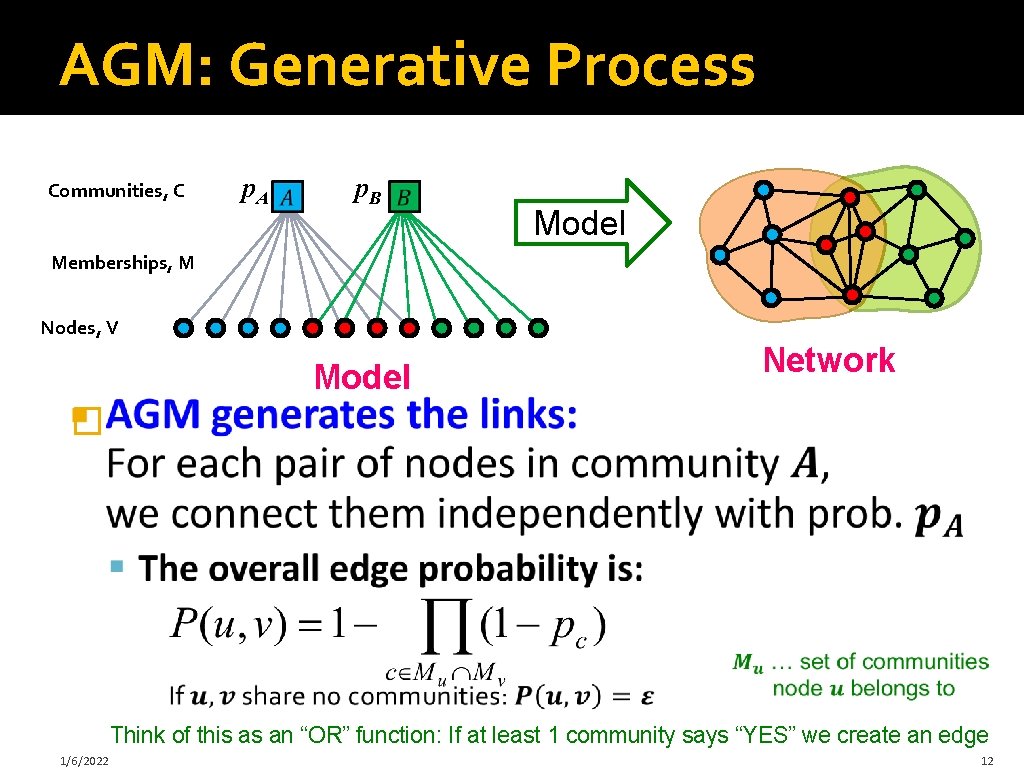
AGM: Generative Process Communities, C p. A p. B Model Memberships, M Nodes, V Model Network � Think of this as an “OR” function: If at least 1 community says “YES” we create an edge 1/6/2022 12
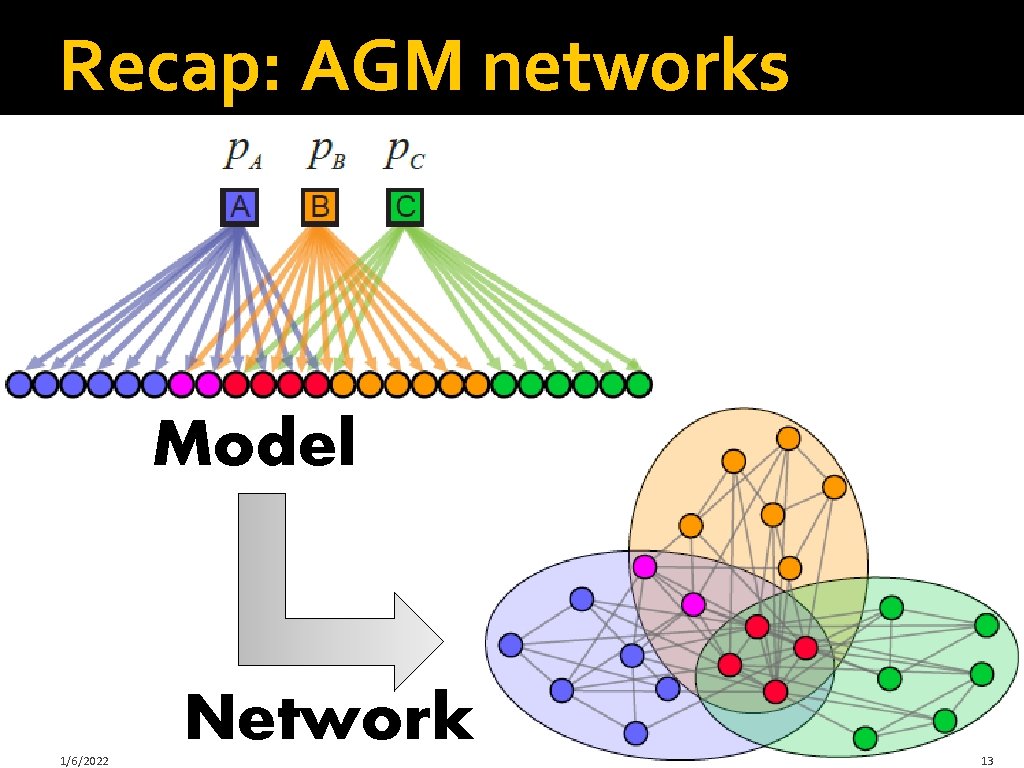
Recap: AGM networks Model 1/6/2022 Network 13
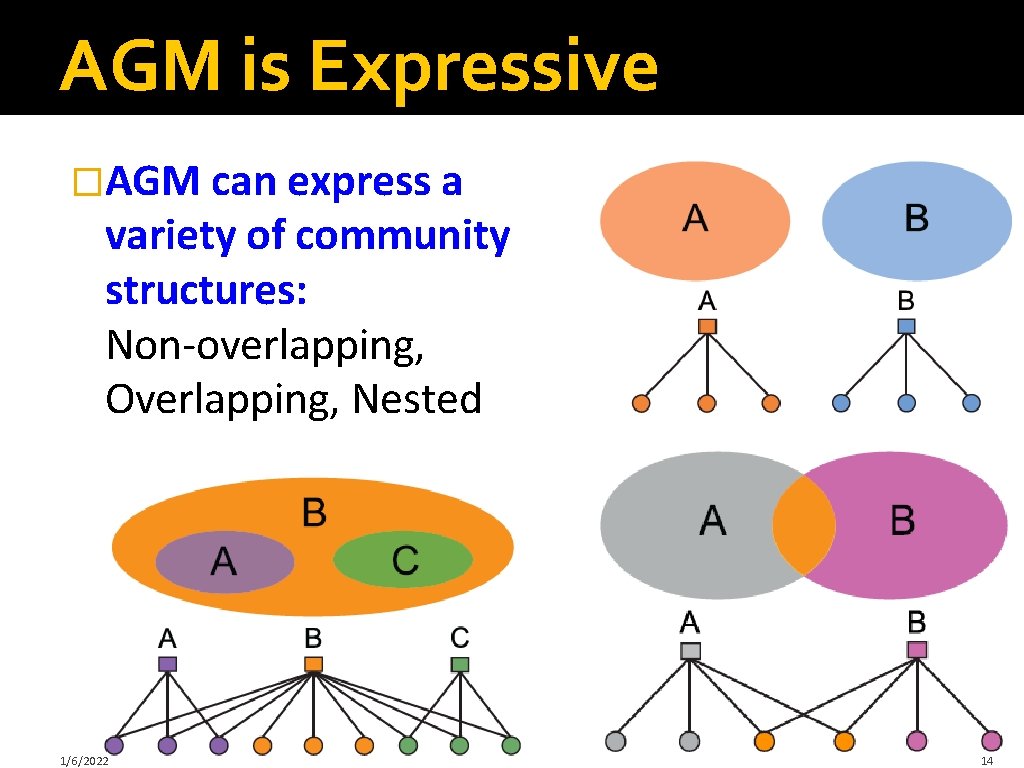
AGM is Expressive �AGM can express a variety of community structures: Non-overlapping, Overlapping, Nested 1/6/2022 14

How do we detect communities with AGM?
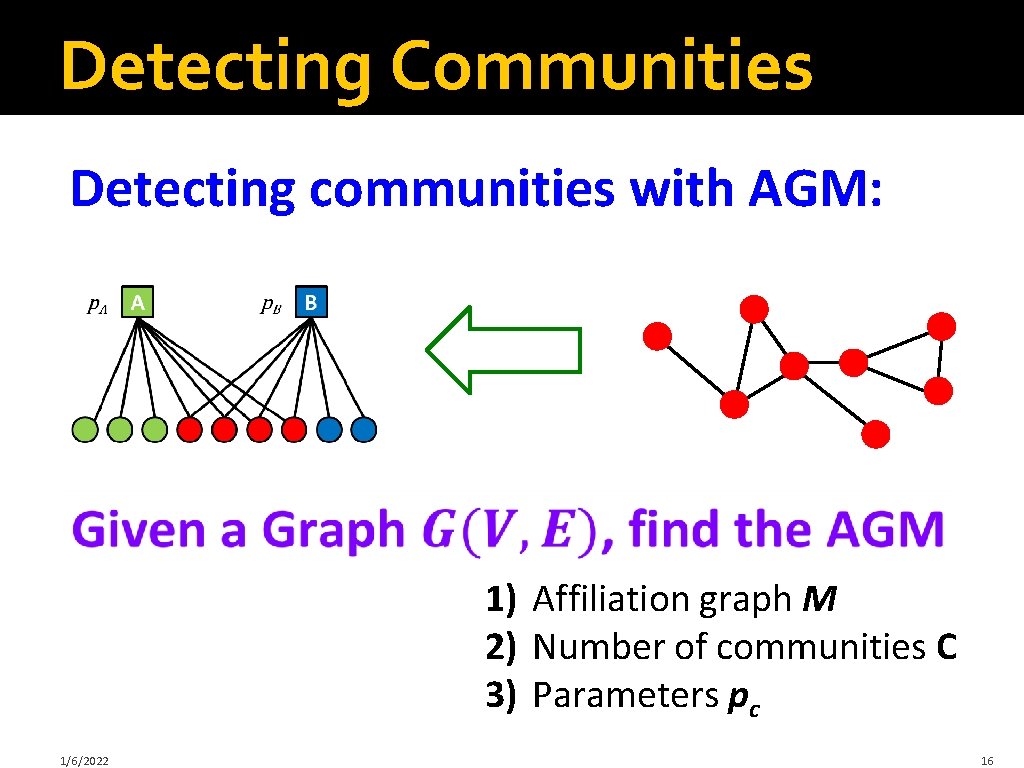
Detecting Communities Detecting communities with AGM: B F A D E G C H 1) Affiliation graph M 2) Number of communities C 3) Parameters pc 1/6/2022 16
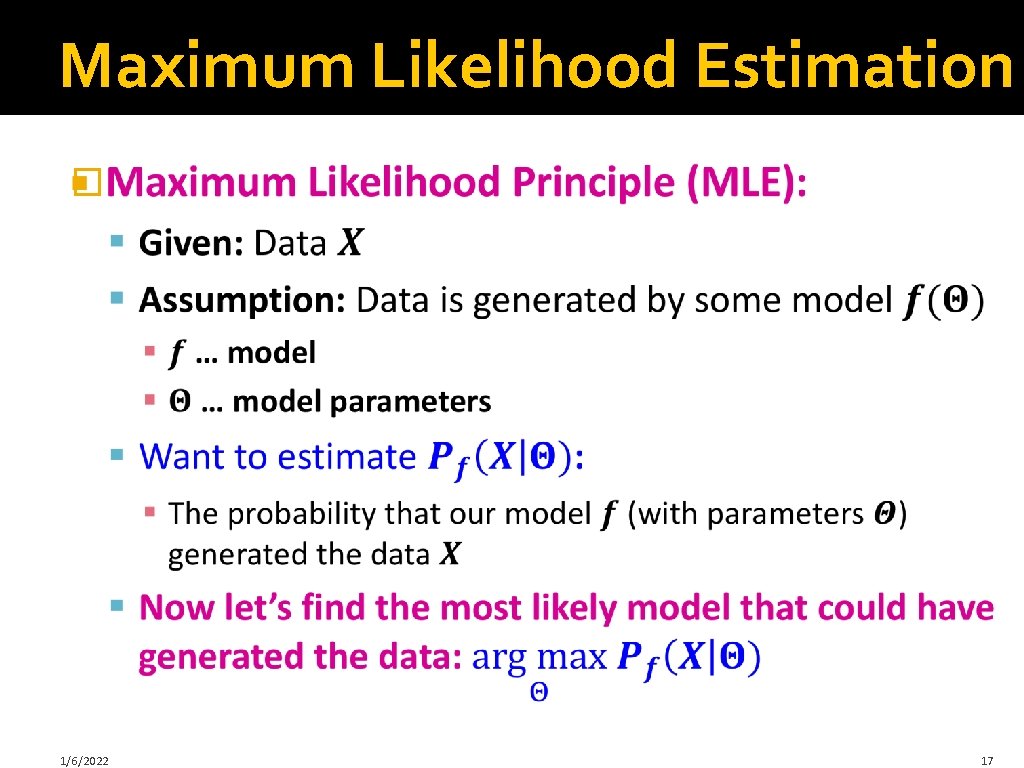
Maximum Likelihood Estimation � 1/6/2022 17
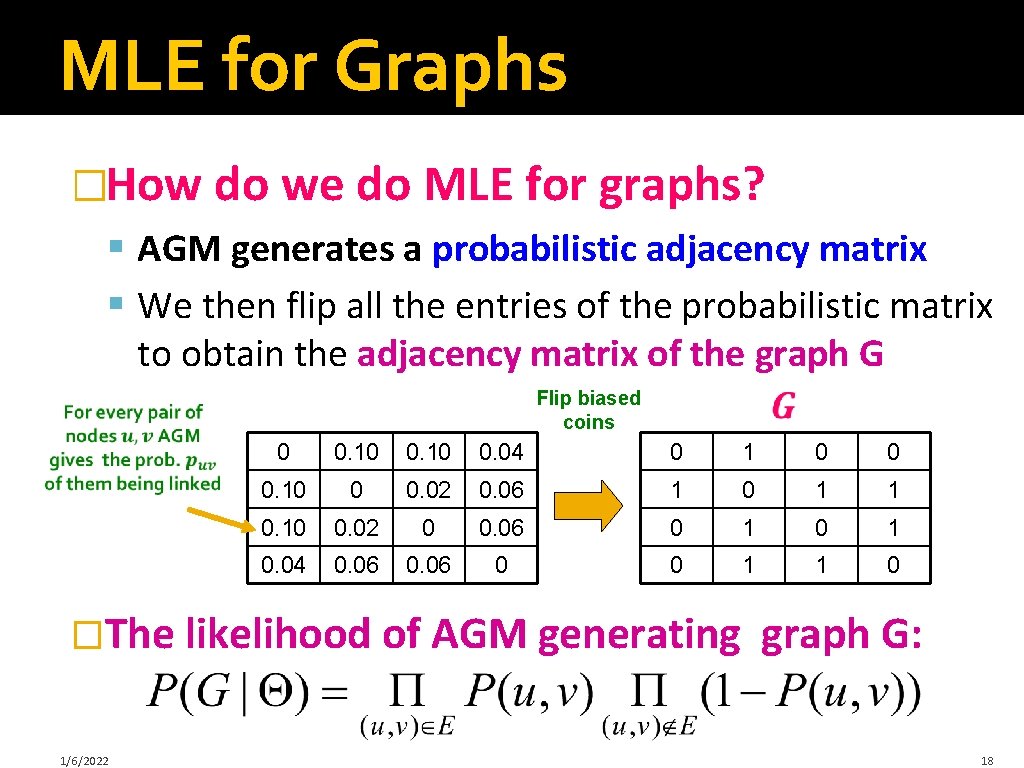
MLE for Graphs �How do we do MLE for graphs? § AGM generates a probabilistic adjacency matrix § We then flip all the entries of the probabilistic matrix to obtain the adjacency matrix of the graph G Flip biased coins 0 0. 10 0. 04 0 1 0 0 0. 10 0 0. 02 0. 06 1 0 1 1 0. 10 0. 02 0 0. 06 0 1 0. 04 0. 06 0 0 1 1 0 �The likelihood of AGM generating 1/6/2022 graph G: 18
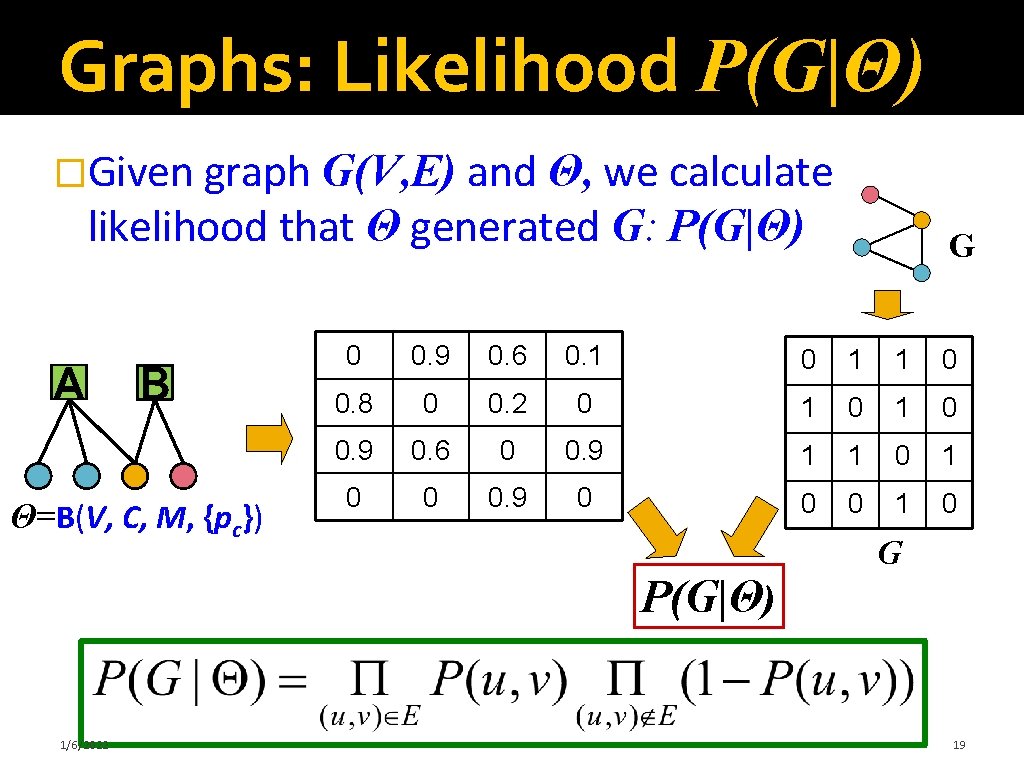
Graphs: Likelihood P(G|Θ) �Given graph G(V, E) and Θ, we calculate likelihood that Θ generated G: P(G|Θ) A B Θ=B(V, C, M, {pc}) G 0 0. 9 0. 6 0. 1 0 1 1 0 0. 8 0 0. 2 0 1 0 0. 9 0. 6 0 0. 9 1 1 0 0 0. 9 0 0 0 1 0 G P(G|Θ) 1/6/2022 19
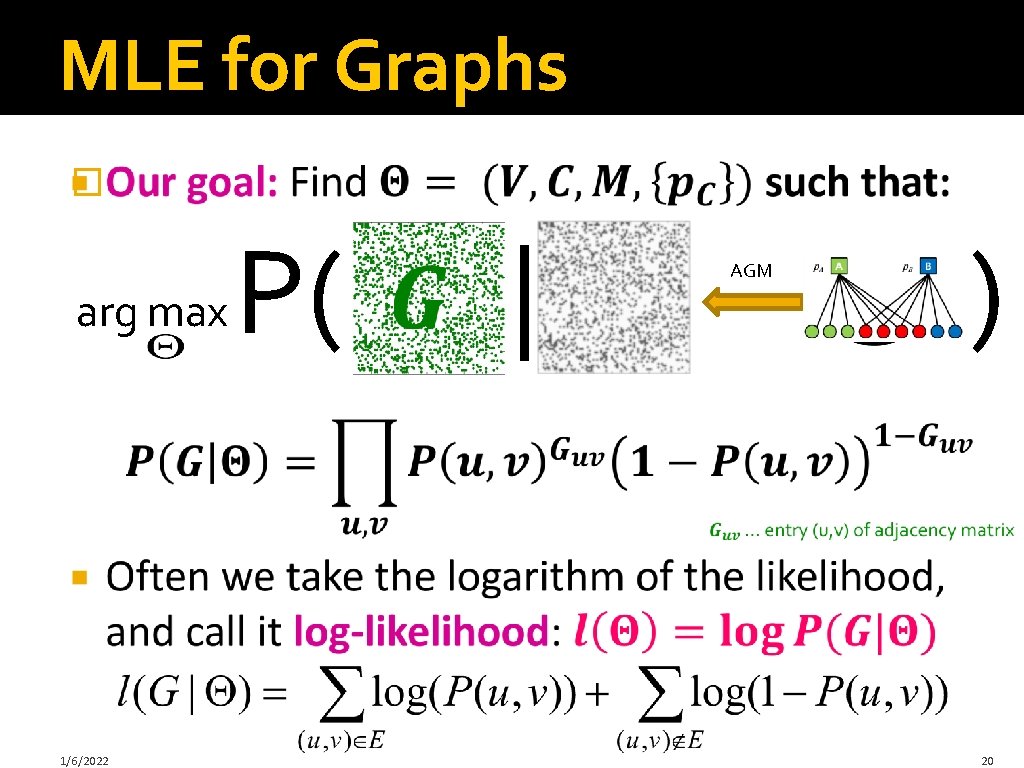
MLE for Graphs � arg max 1/6/2022 P( | AGM ) 20
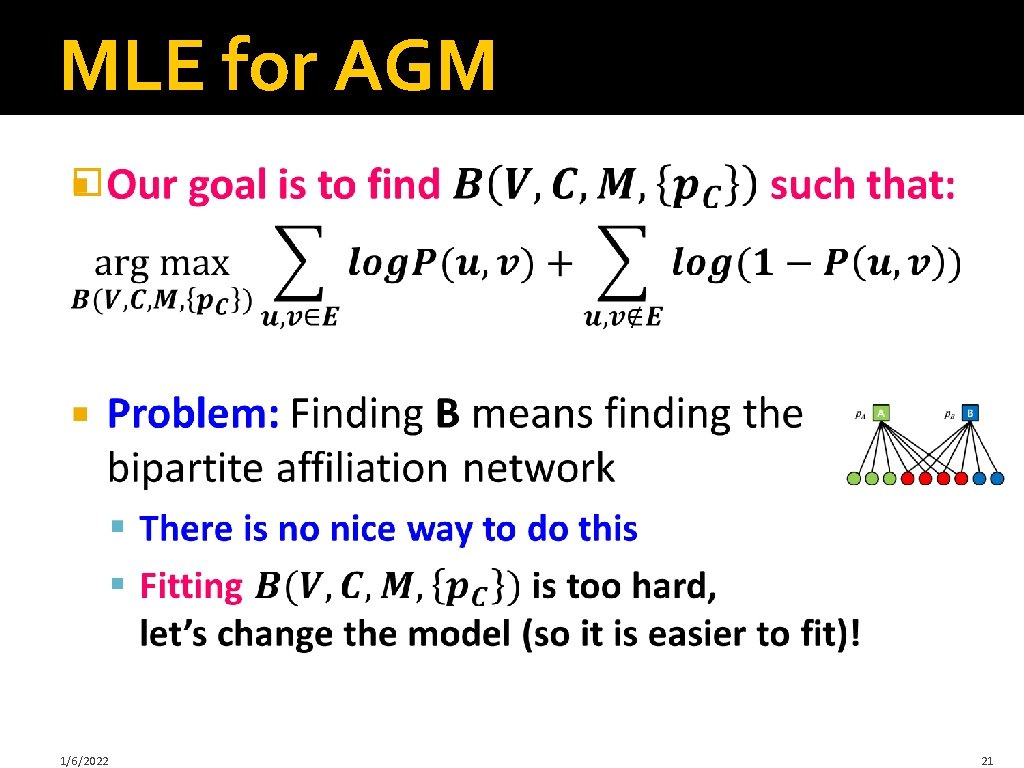
MLE for AGM � 1/6/2022 21
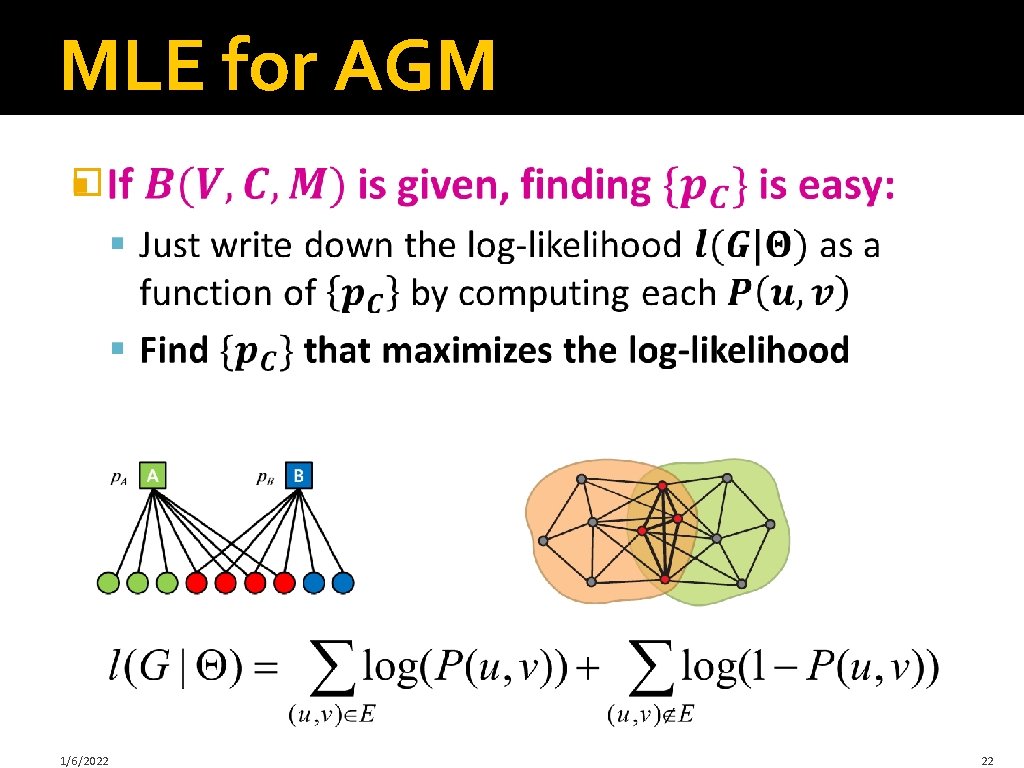
MLE for AGM � 1/6/2022 22
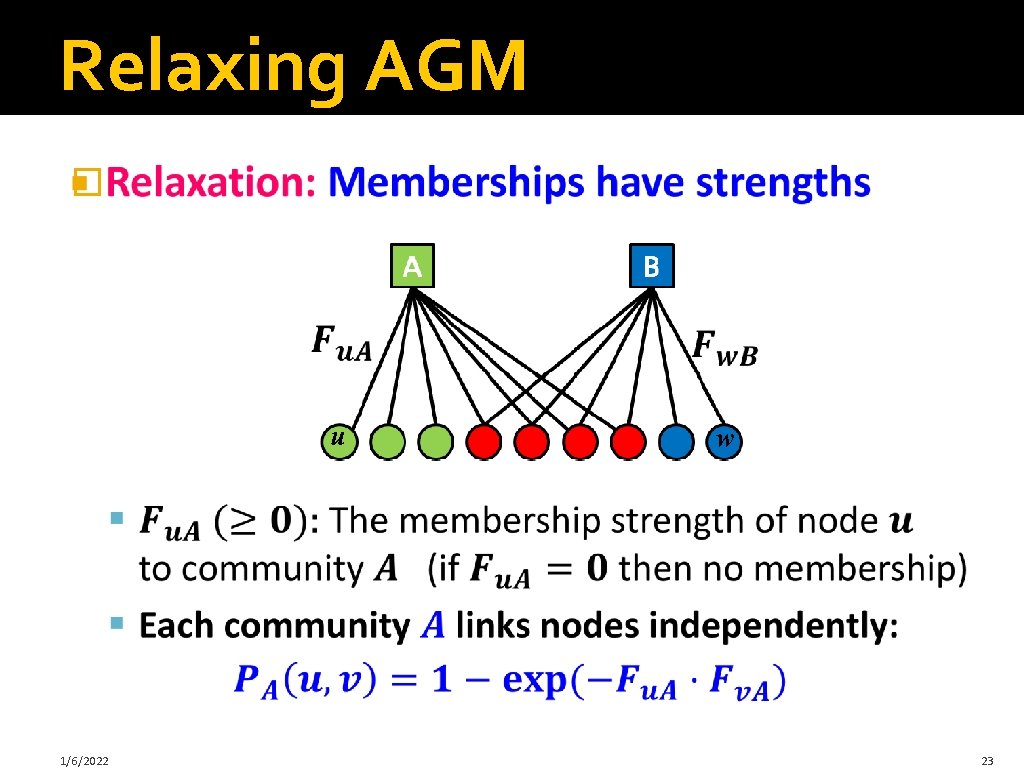
Relaxing AGM � u 1/6/2022 w 23
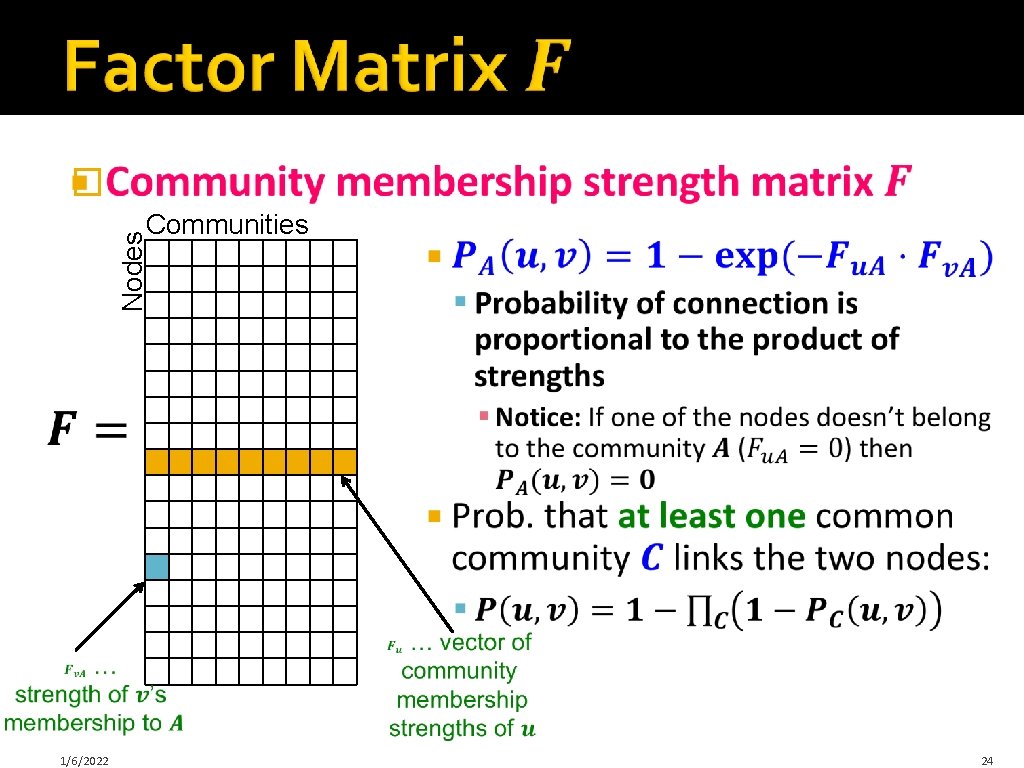
� Nodes Communities j 1/6/2022 24
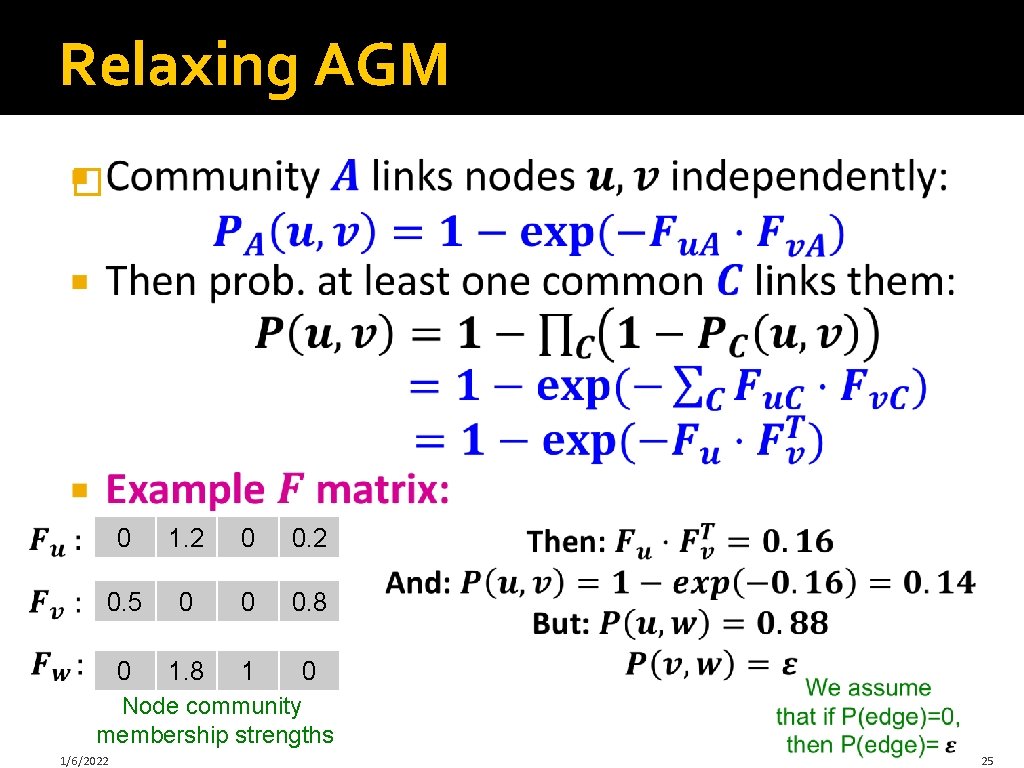
Relaxing AGM � 0 1. 2 0 0. 2 0. 5 0 0 0. 8 0 1. 8 1 0 Node community membership strengths 1/6/2022 25
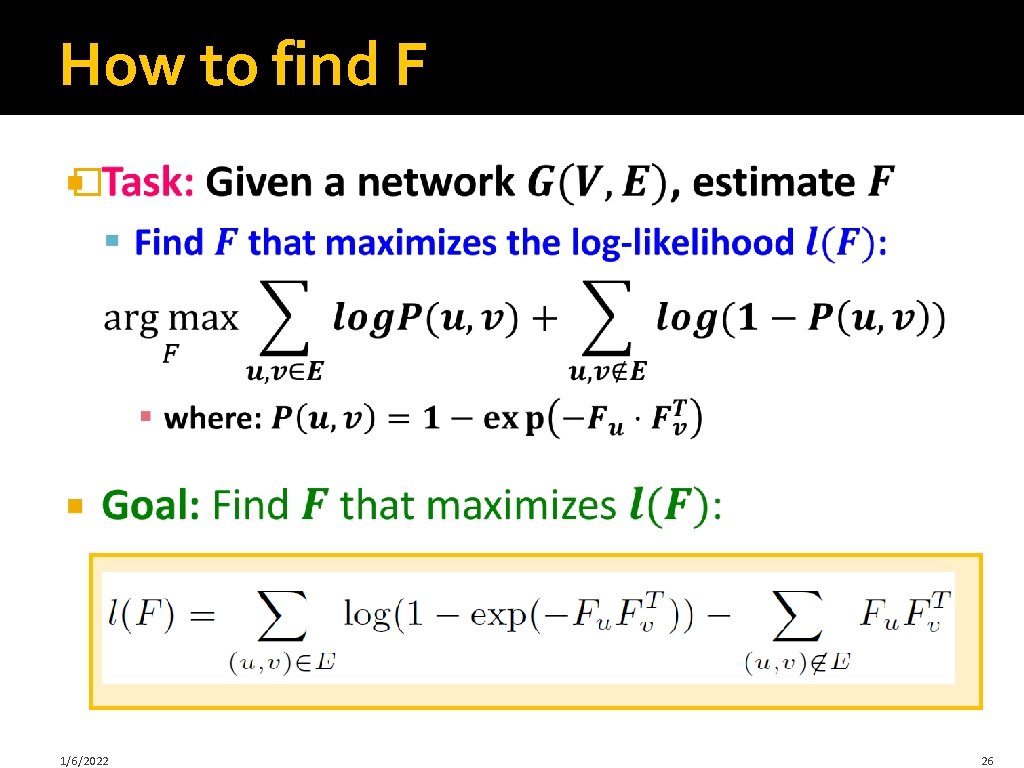
How to find F � 1/6/2022 26
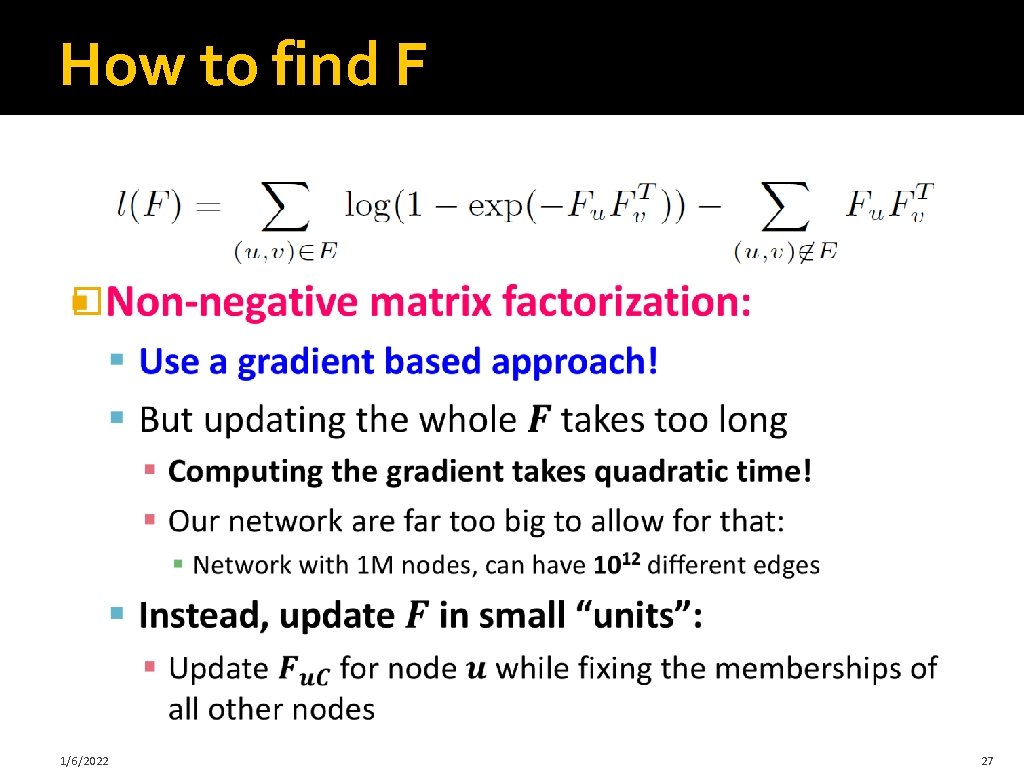
How to find F � 1/6/2022 27
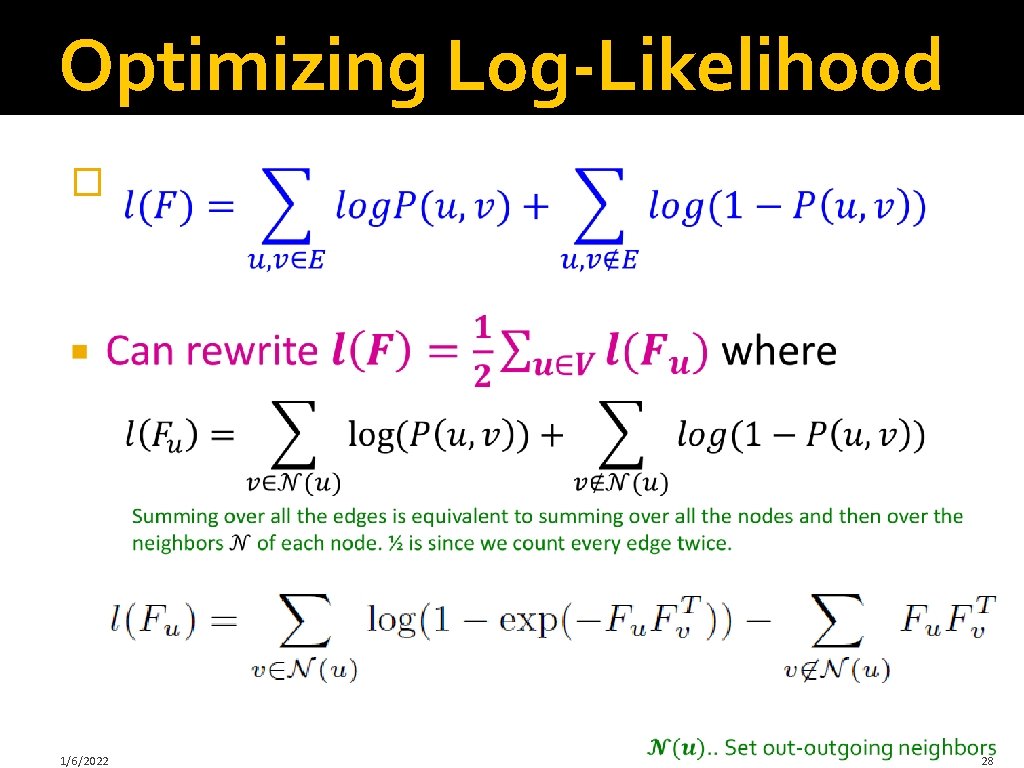
Optimizing Log-Likelihood � 1/6/2022 28
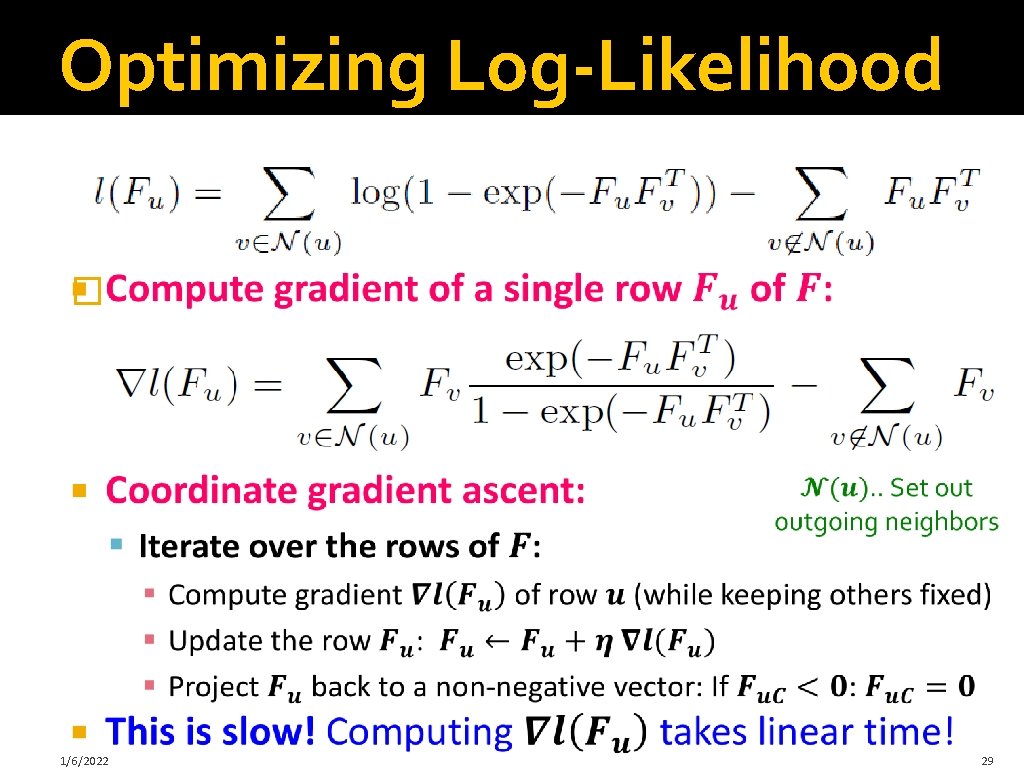
Optimizing Log-Likelihood � 1/6/2022 29
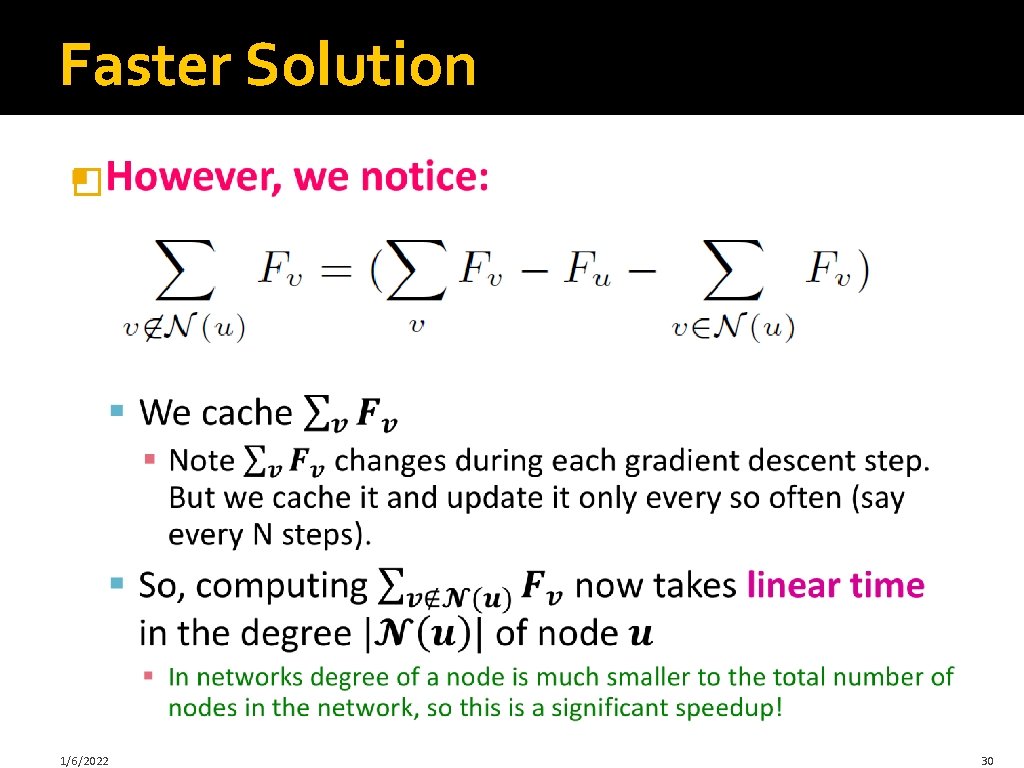
Faster Solution � 1/6/2022 30

Node Classification in Networks: Guilt by Association
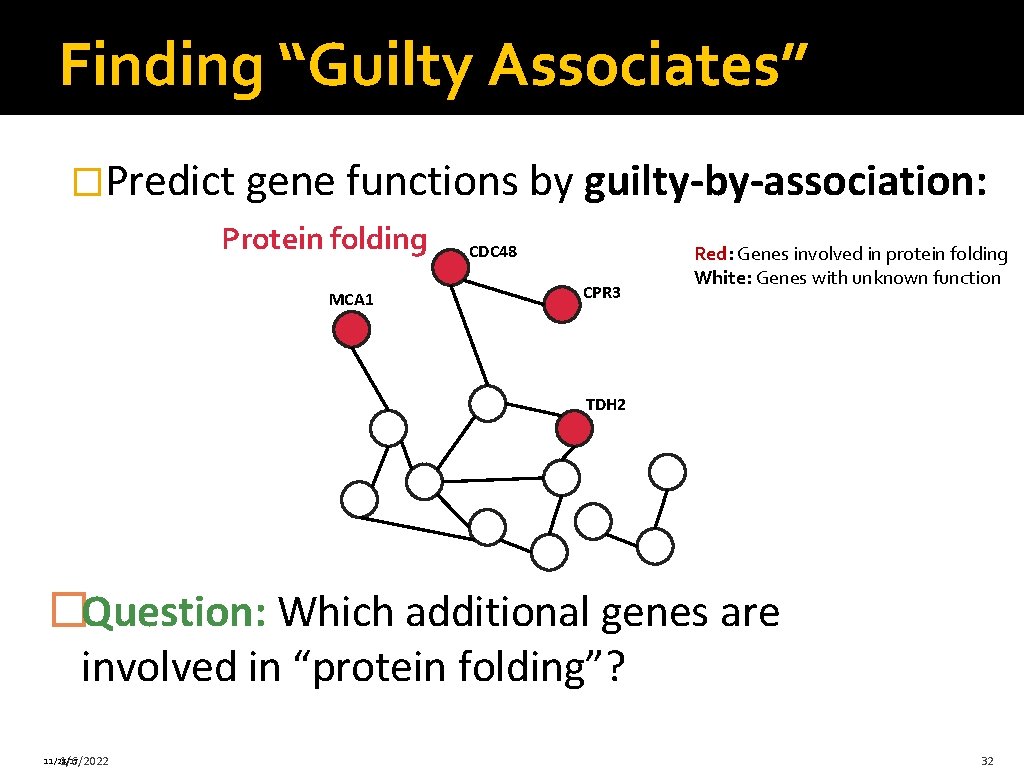
Finding “Guilty Associates” �Predict gene functions by guilty-by-association: Protein folding MCA 1 CDC 48 CPR 3 Red: Genes involved in protein folding White: Genes with unknown function TDH 2 �Question: Which additional genes are involved in “protein folding”? 11/28/17 1/6/2022 32

“Guilty Associates” Problem � 11/28/17 1/6/2022 33
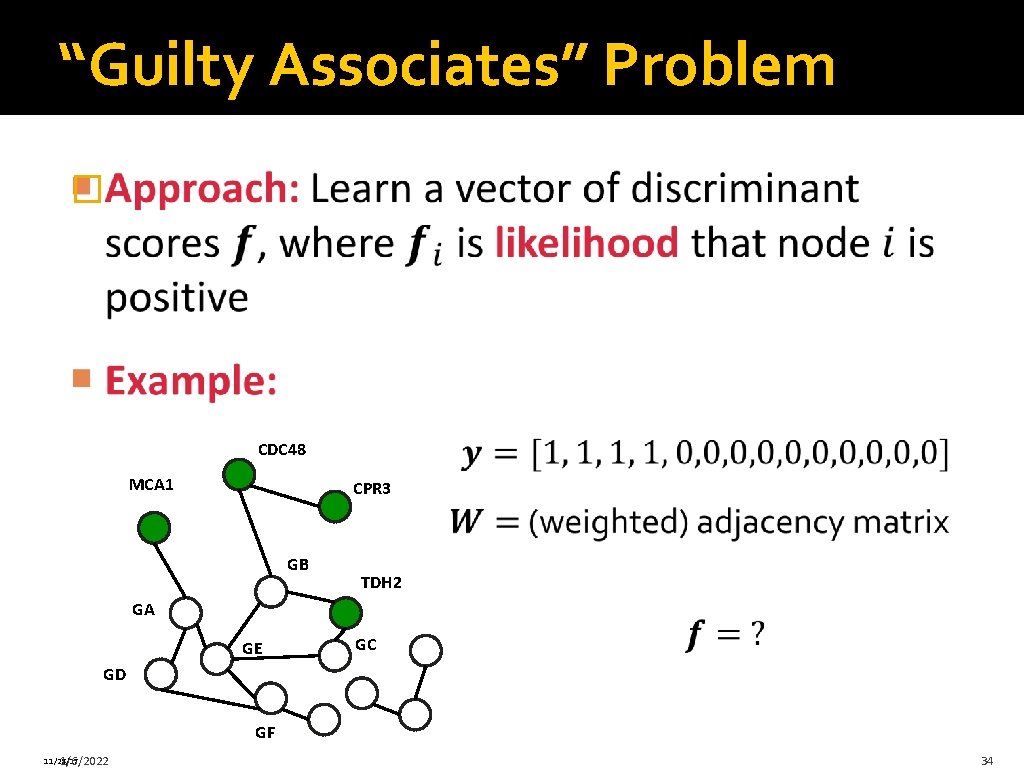
“Guilty Associates” Problem � CDC 48 MCA 1 CPR 3 GB TDH 2 GA GE GC GD GF 11/28/17 1/6/2022 34

Three Approaches Three approaches to Guilt by Association � 1) Neighbor scoring � 2) Using Random Walks � 3) Label propagation 1/6/2022 35
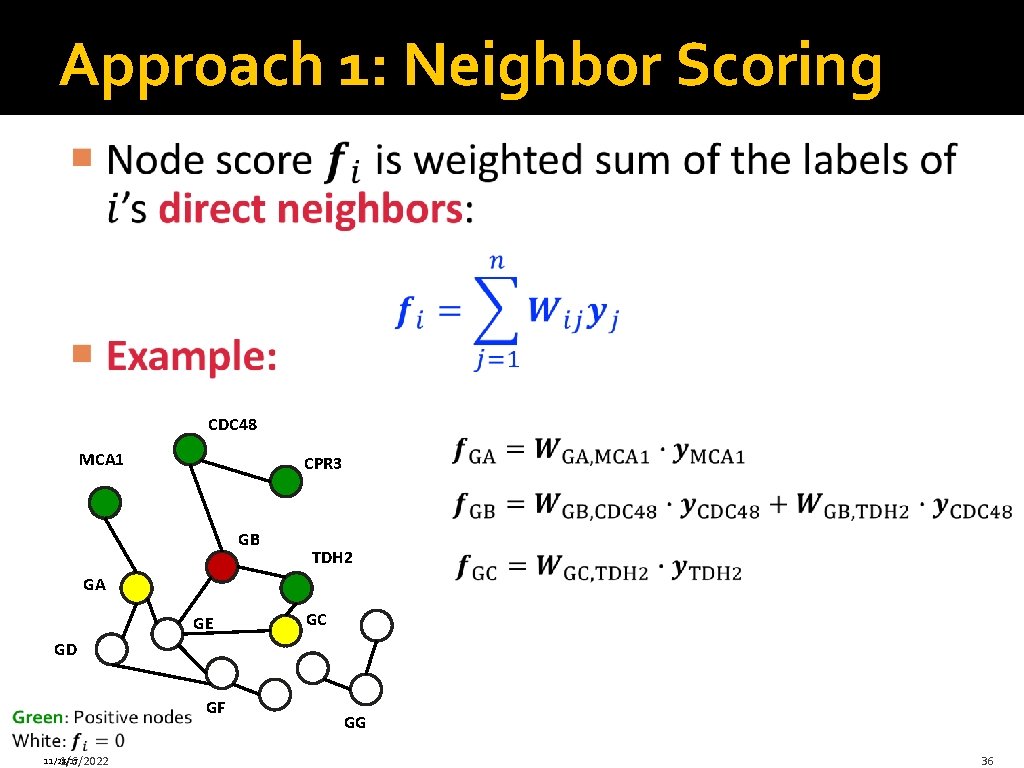
Approach 1: Neighbor Scoring CDC 48 MCA 1 CPR 3 GB TDH 2 GA GE GC GD GF 11/28/17 1/6/2022 GG 36
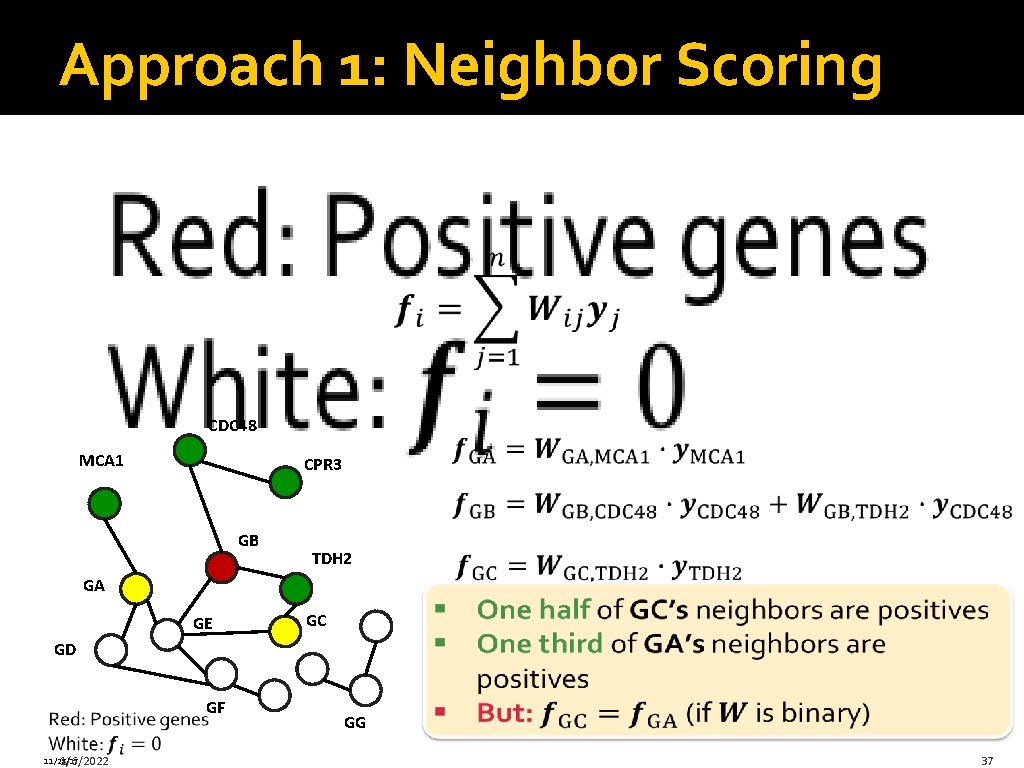
Approach 1: Neighbor Scoring CDC 48 MCA 1 CPR 3 GB TDH 2 GA GE GC GD GF 11/28/17 1/6/2022 GG 37
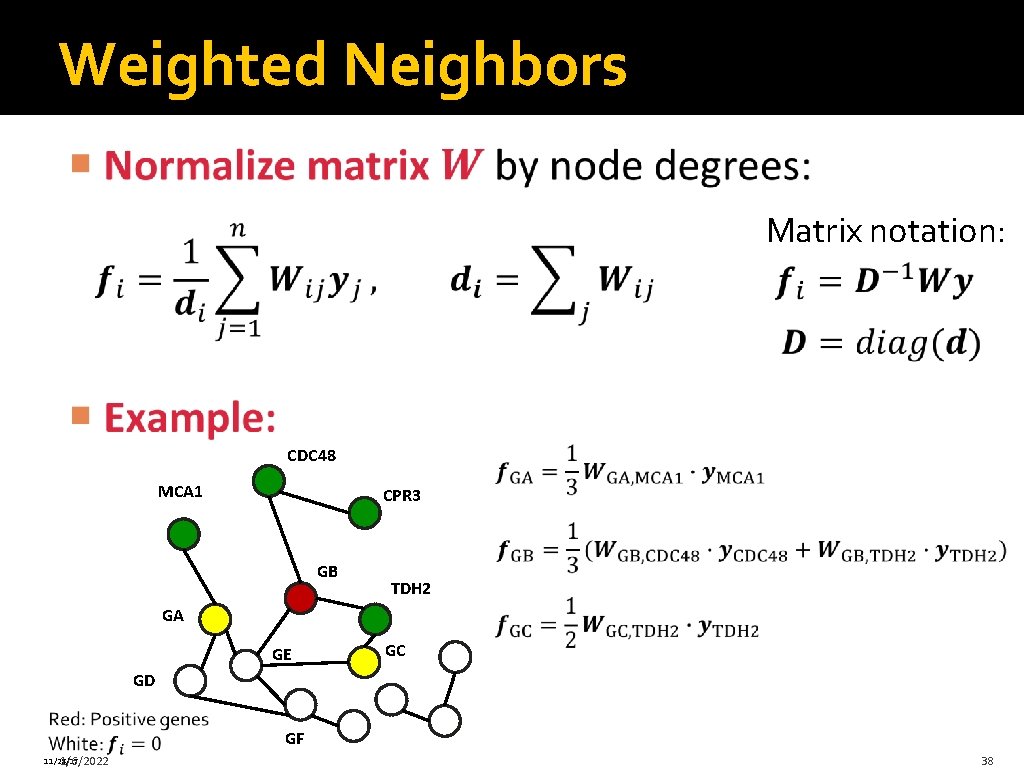
Weighted Neighbors Matrix notation: CDC 48 MCA 1 CPR 3 GB TDH 2 GA GE GC GD GF 11/28/17 1/6/2022 38

Indirect Neighbor Scoring � 11/28/17 1/6/2022 39
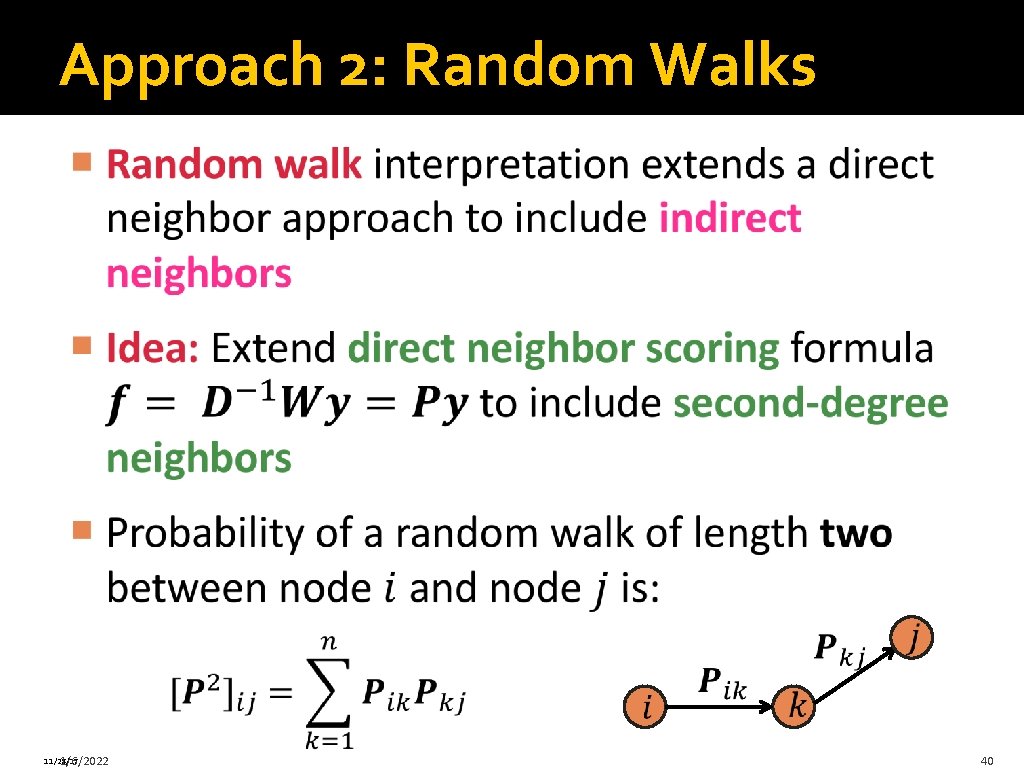
Approach 2: Random Walks 11/28/17 1/6/2022 40
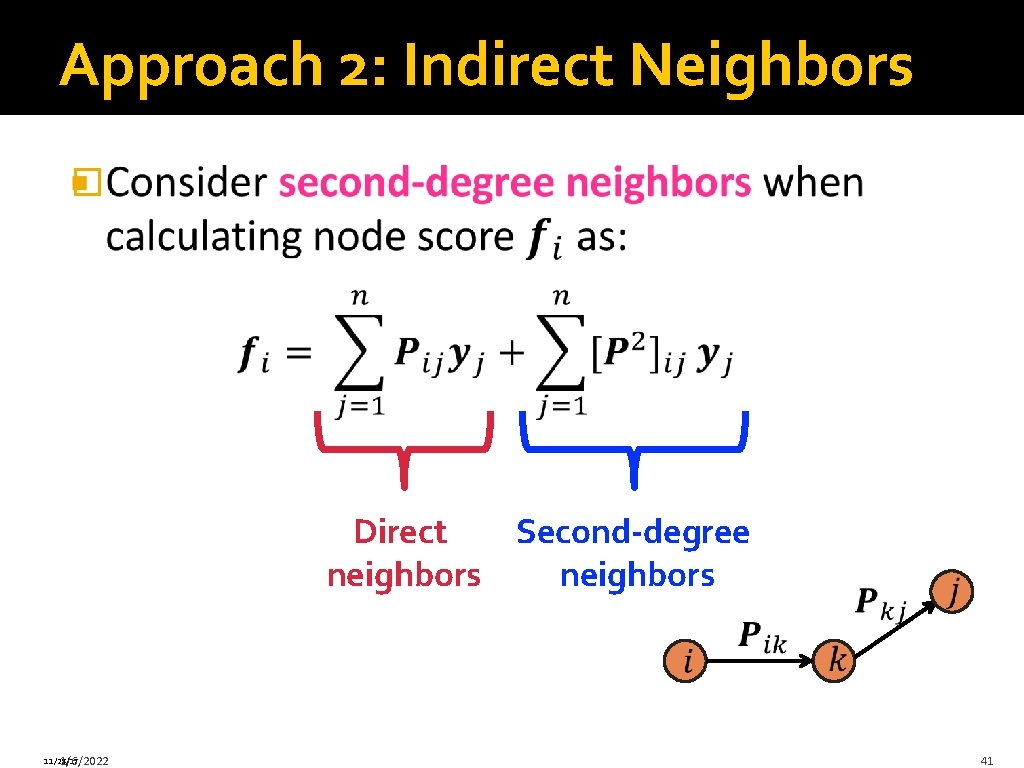
Approach 2: Indirect Neighbors � Direct Second-degree neighbors 11/28/17 1/6/2022 41
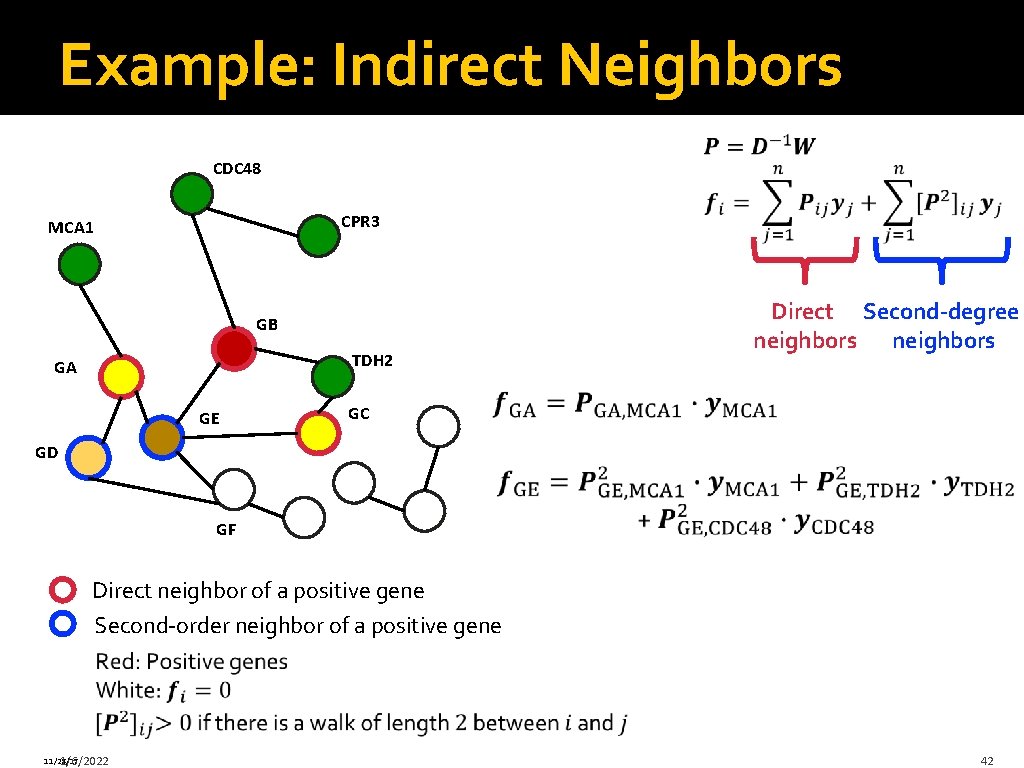
Example: Indirect Neighbors CDC 48 CPR 3 MCA 1 GB TDH 2 GA GE Direct Second-degree neighbors GC GD GF Direct neighbor of a positive gene Second-order neighbor of a positive gene 11/28/17 1/6/2022 42

Beyond Second-Degree Neighbors � 1/6/2022 Jure Leskovec, Stanford CS 246: Mining Massive Datasets 43
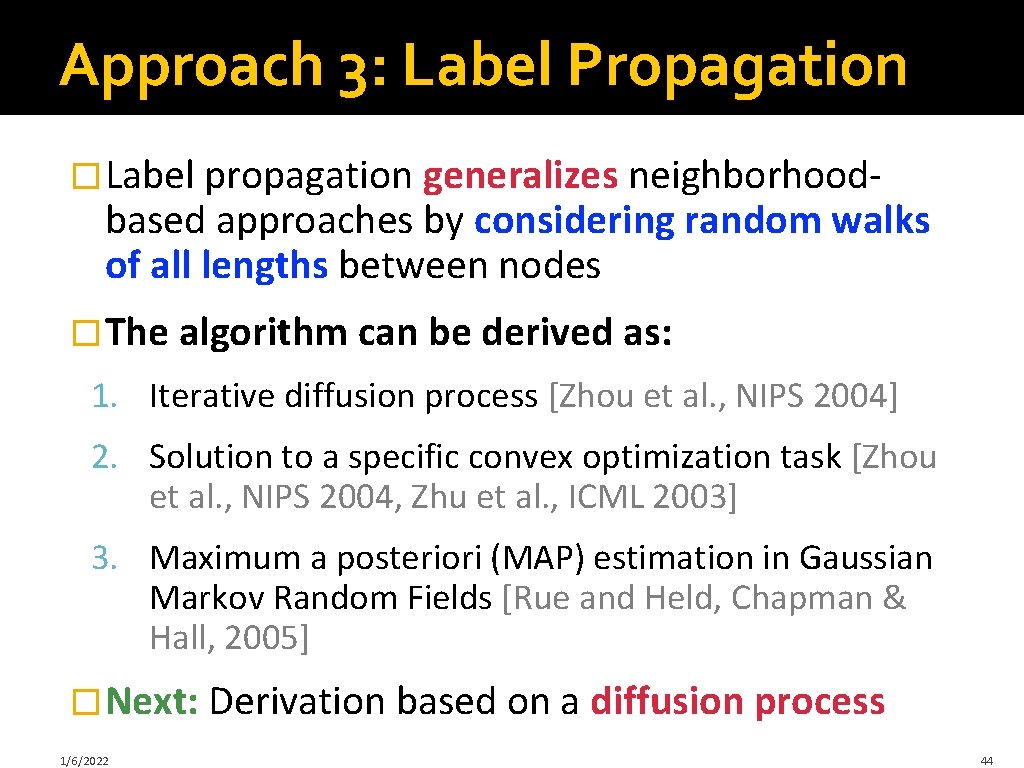
Approach 3: Label Propagation � Label propagation generalizes neighborhood- based approaches by considering random walks of all lengths between nodes � The algorithm can be derived as: 1. Iterative diffusion process [Zhou et al. , NIPS 2004] 2. Solution to a specific convex optimization task [Zhou et al. , NIPS 2004, Zhu et al. , ICML 2003] 3. Maximum a posteriori (MAP) estimation in Gaussian Markov Random Fields [Rue and Held, Chapman & Hall, 2005] � Next: Derivation based on a diffusion process 1/6/2022 44
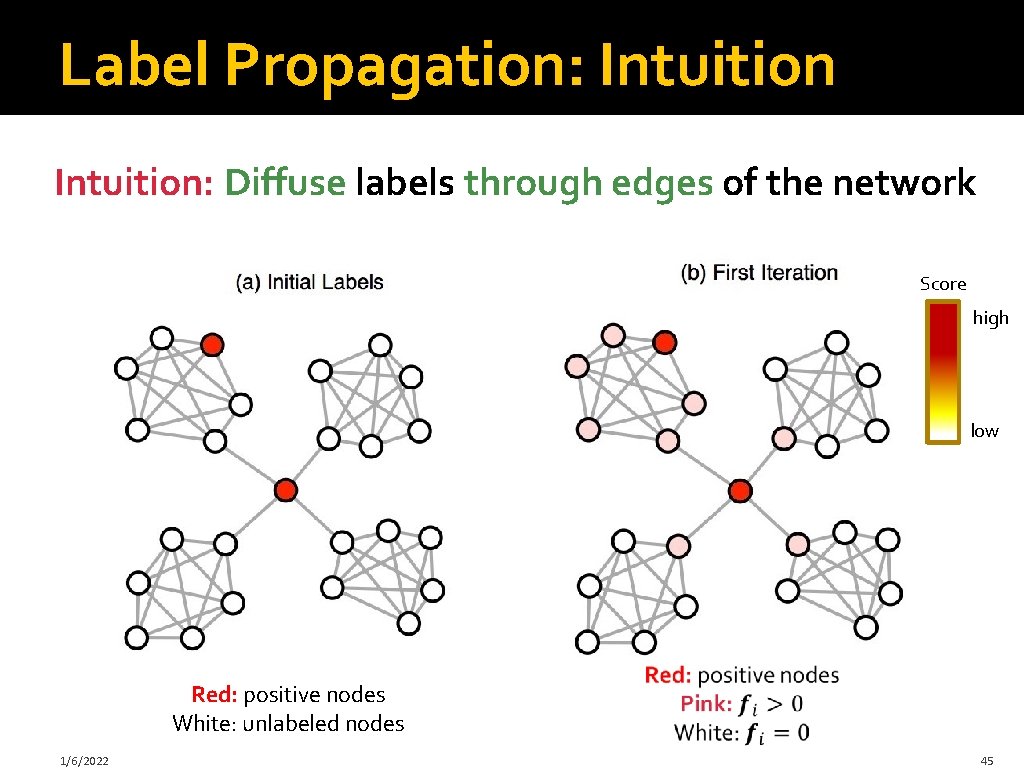
Label Propagation: Intuition: Diffuse labels through edges of the network Score high low Red: positive nodes White: unlabeled nodes 1/6/2022 45
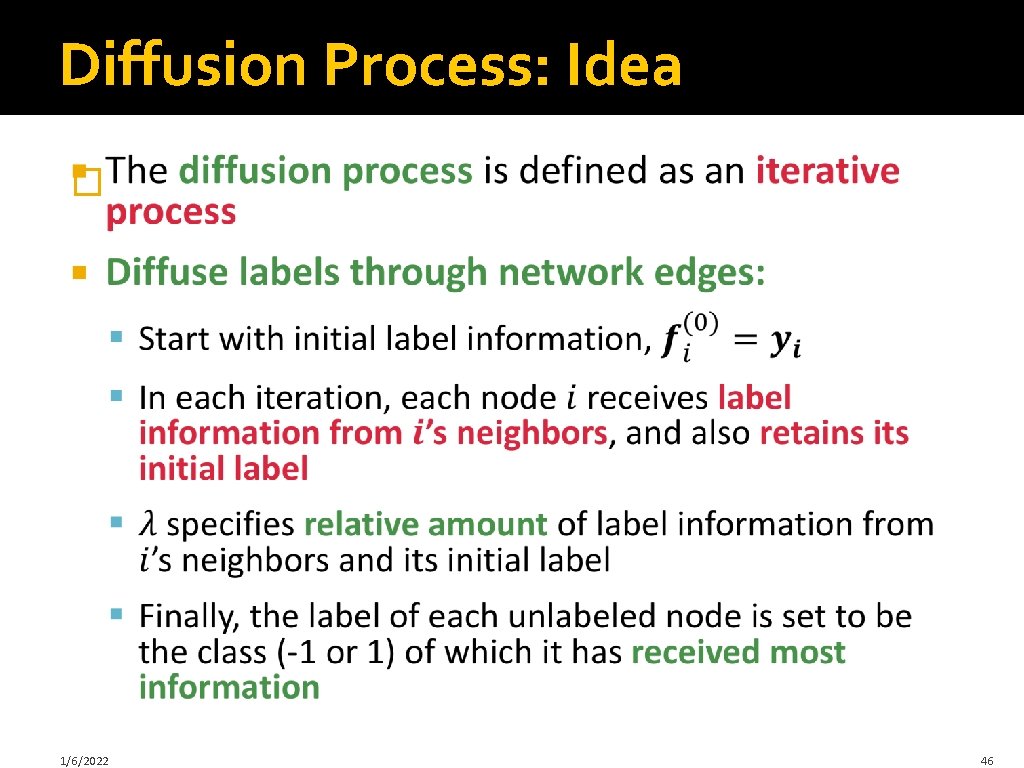
Diffusion Process: Idea � 1/6/2022 46
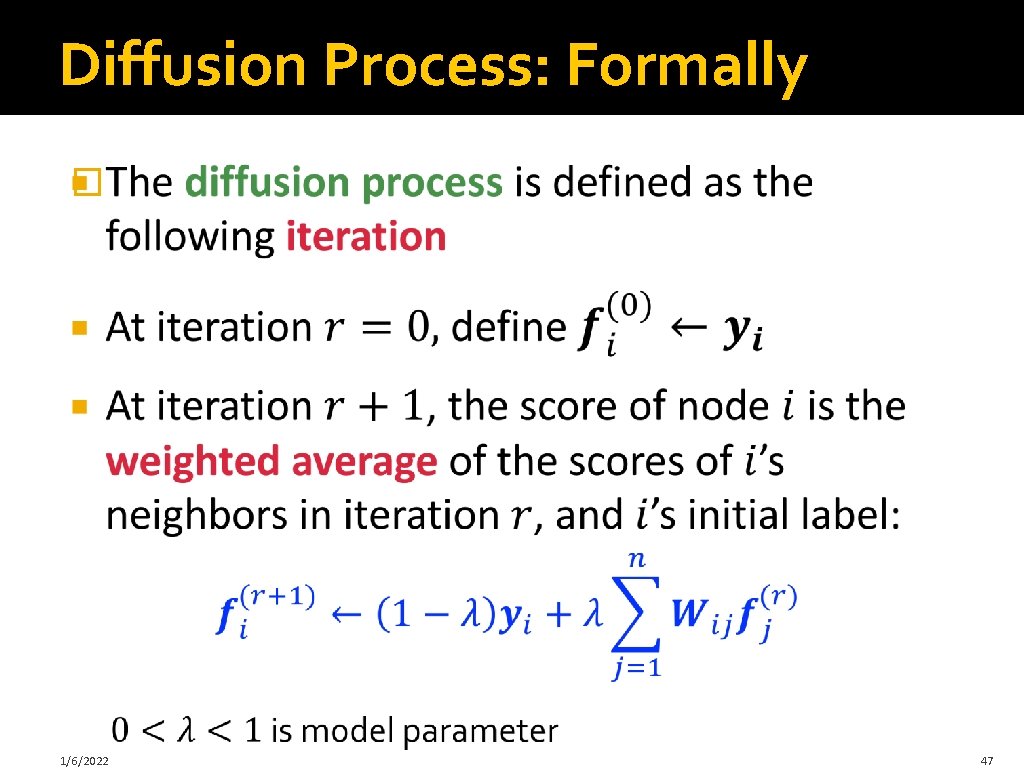
Diffusion Process: Formally � 1/6/2022 47
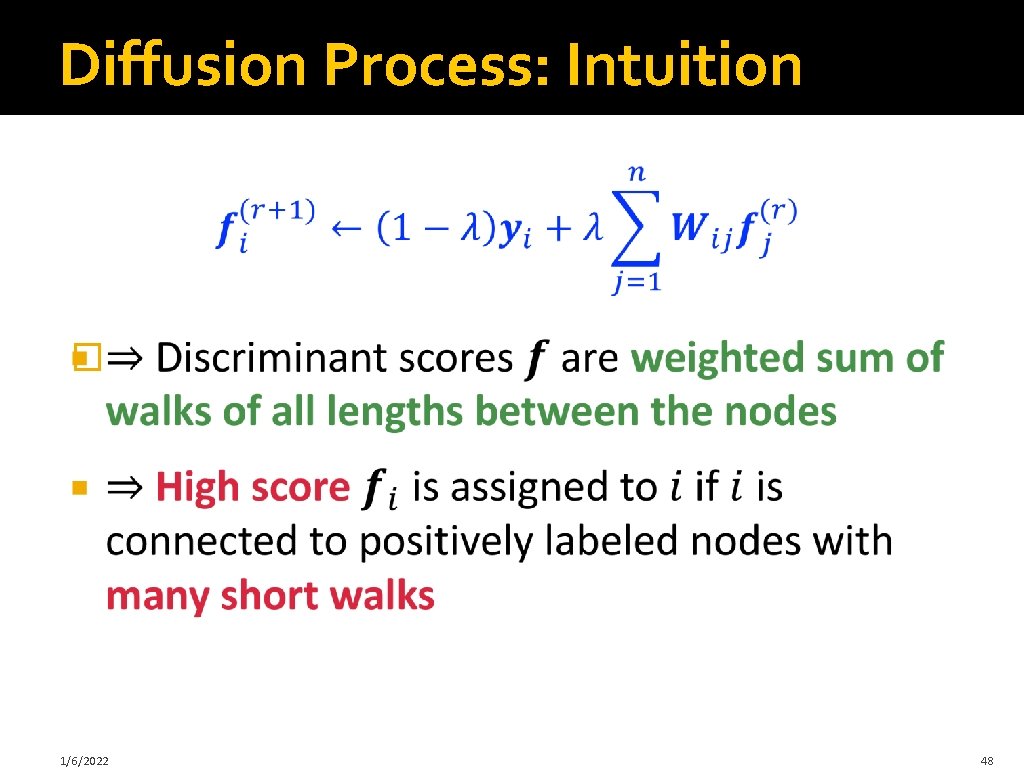
Diffusion Process: Intuition � 1/6/2022 48
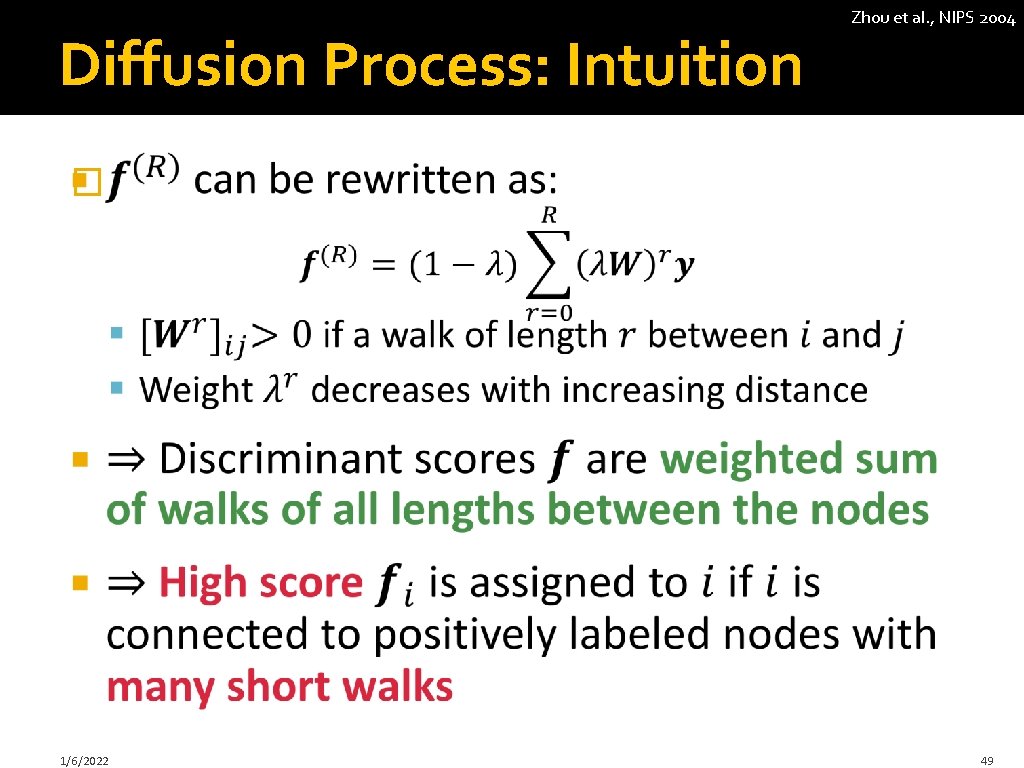
Diffusion Process: Intuition Zhou et al. , NIPS 2004 � 1/6/2022 49
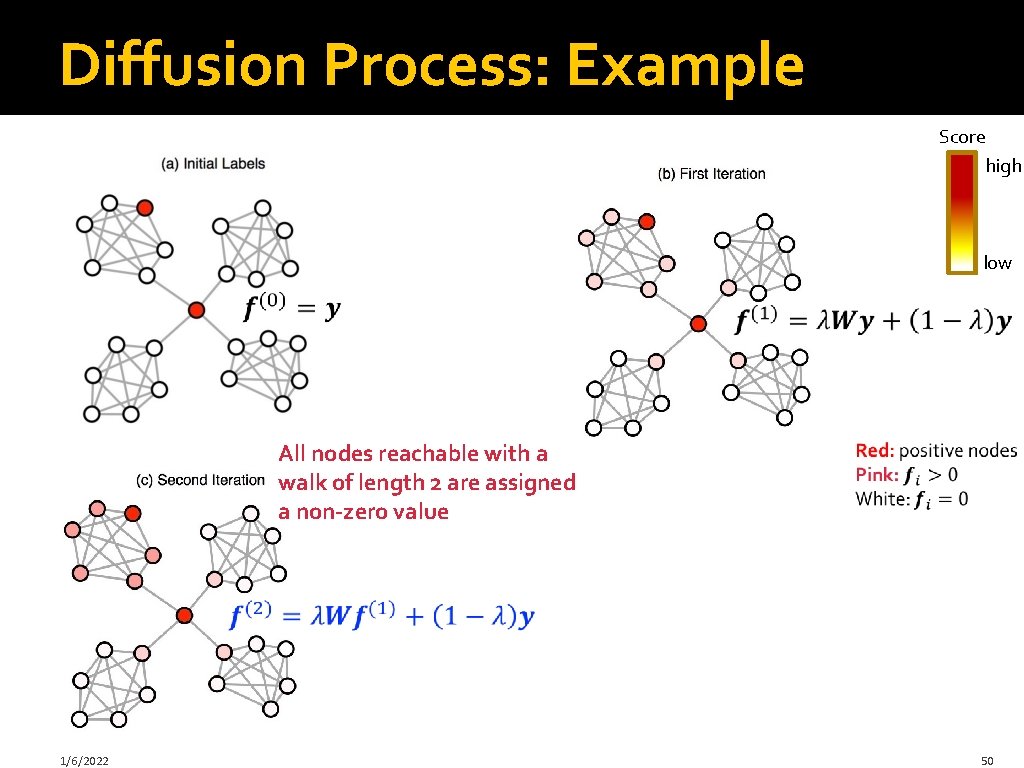
Diffusion Process: Example Score high low All nodes reachable with a walk of length 2 are assigned a non-zero value 1/6/2022 50
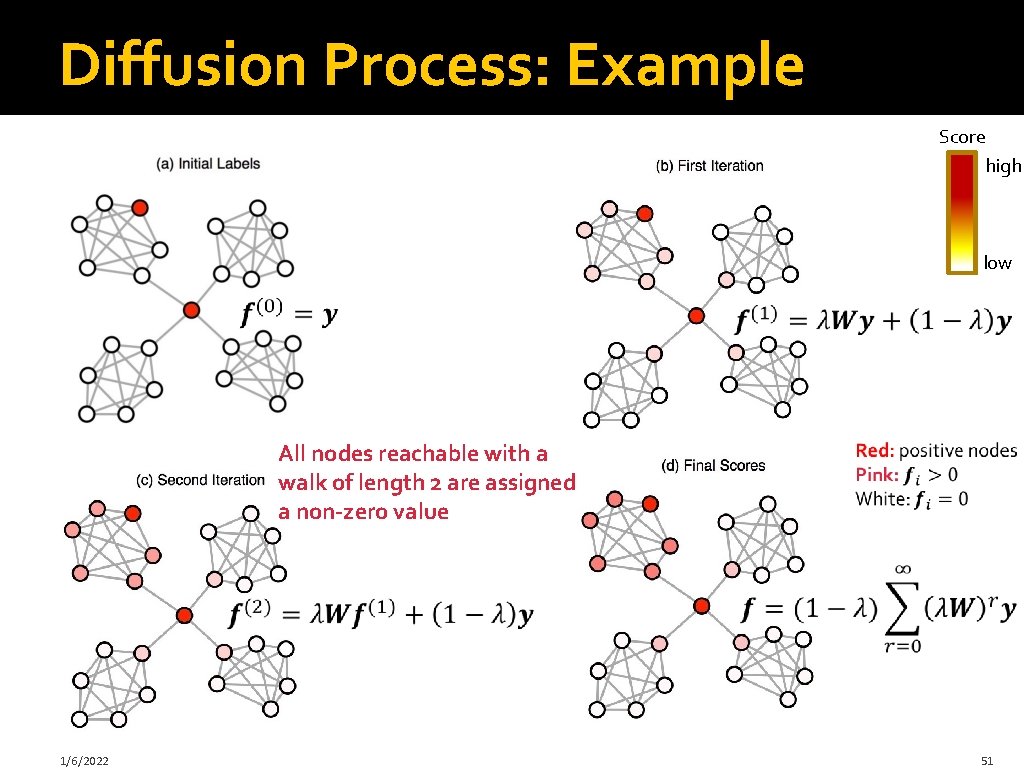
Diffusion Process: Example Score high low All nodes reachable with a walk of length 2 are assigned a non-zero value 1/6/2022 51
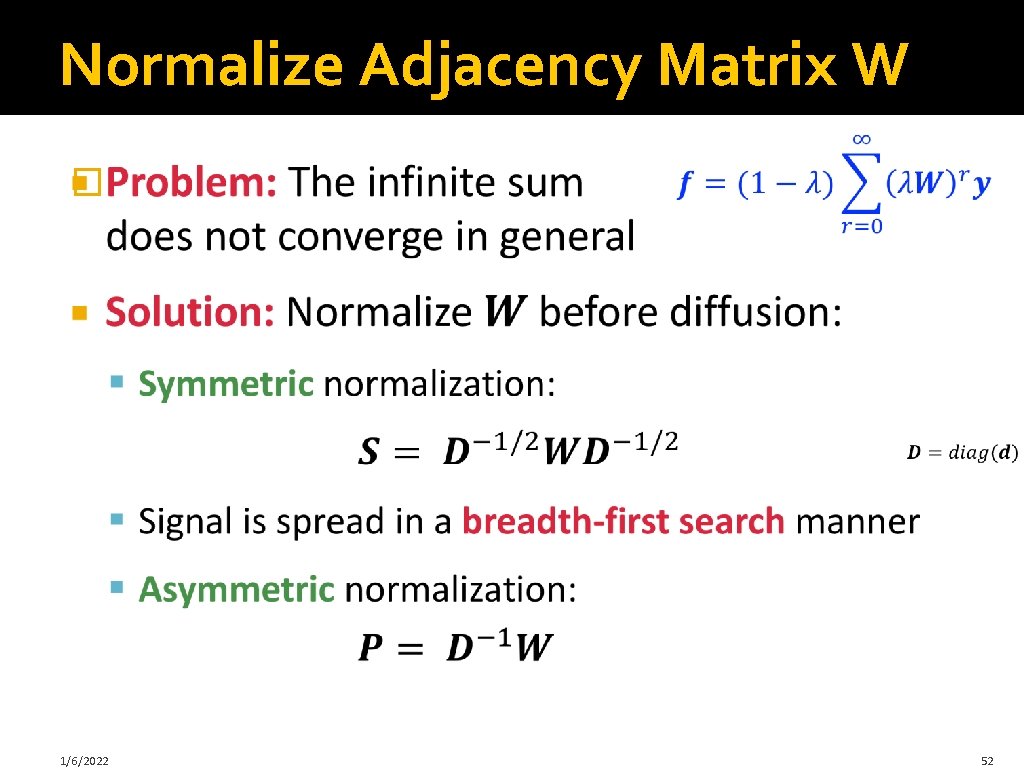
Normalize Adjacency Matrix W � 1/6/2022 52
Function Prediction: Setup � 1/6/2022 53
- Slides: 53