CNCP 2012 Beijing Can Pro Var 2 0

第二届中国计算蛋白质组学研讨会 CNCP 2012, Beijing Can. Pro. Var 2. 0 : Updated Database of Human Cancer Proteome Variation Jing Li jing. li@sjtu. edu. cn CNCP 2012 November 2012
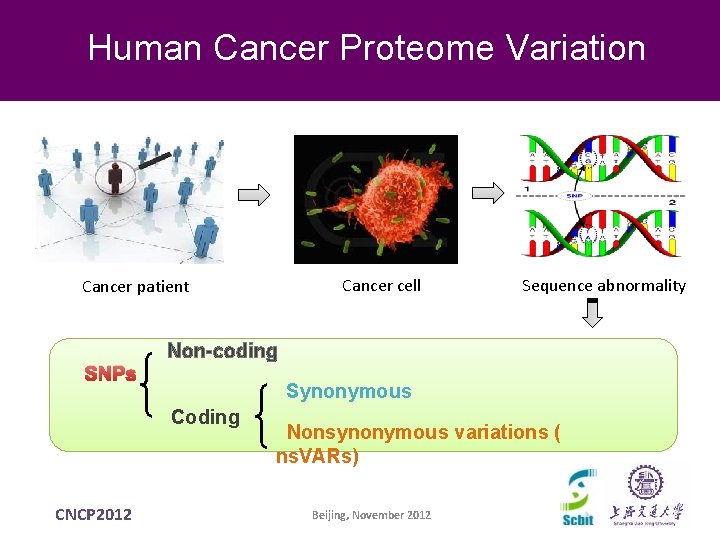
Human Cancer Proteome Variation Cancer cell Cancer patient SNPs Non-coding Synonymous Coding CNCP 2012 Sequence abnormality Nonsynonymous variations ( ns. VARs) Beijing, November 2012
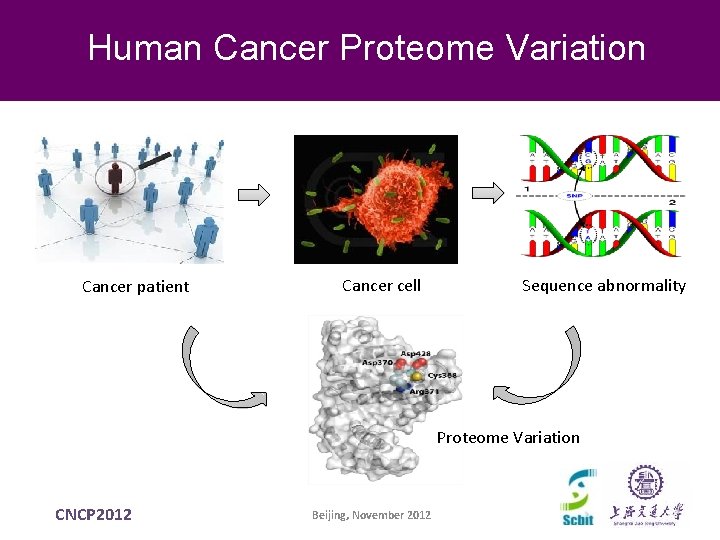
Human Cancer Proteome Variation Cancer patient Cancer cell Sequence abnormality Proteome Variation CNCP 2012 Beijing, November 2012
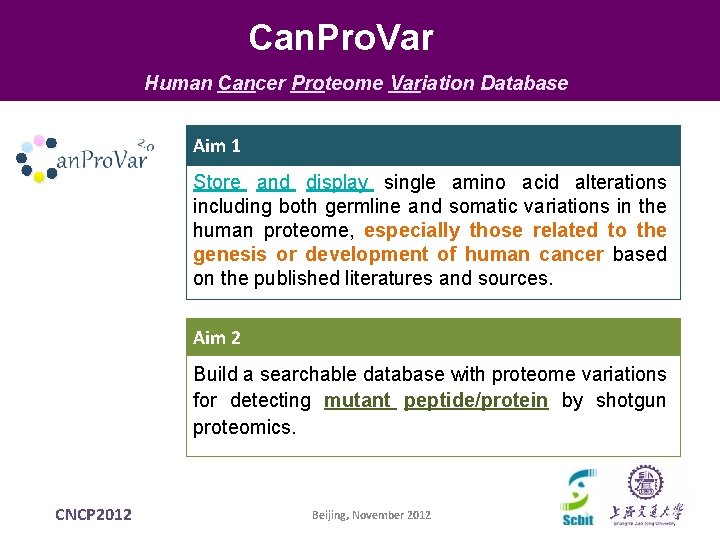
Can. Pro. Var Human Cancer Proteome Variation Database Aim 1 Store and display single amino acid alterations including both germline and somatic variations in the human proteome, especially those related to the genesis or development of human cancer based on the published literatures and sources. Aim 2 Build a searchable database with proteome variations for detecting mutant peptide/protein by shotgun proteomics. CNCP 2012 Beijing, November 2012
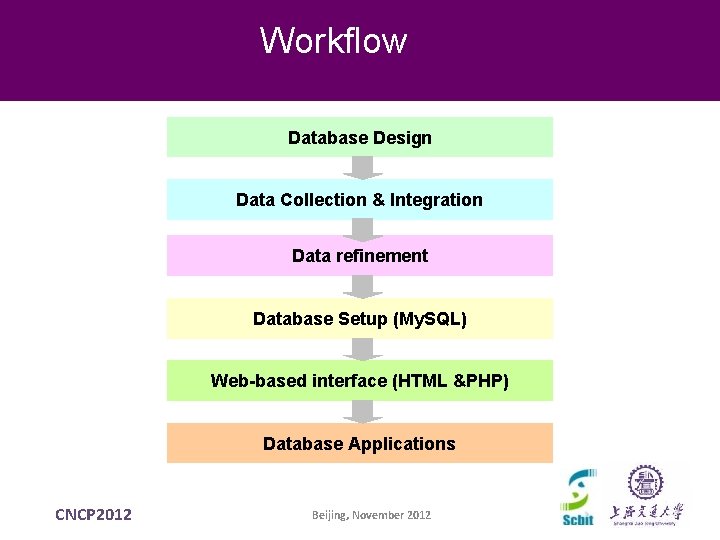
Workflow Database Design Data Collection & Integration Data refinement Database Setup (My. SQL) Web-based interface (HTML &PHP) Database Applications CNCP 2012 Beijing, November 2012
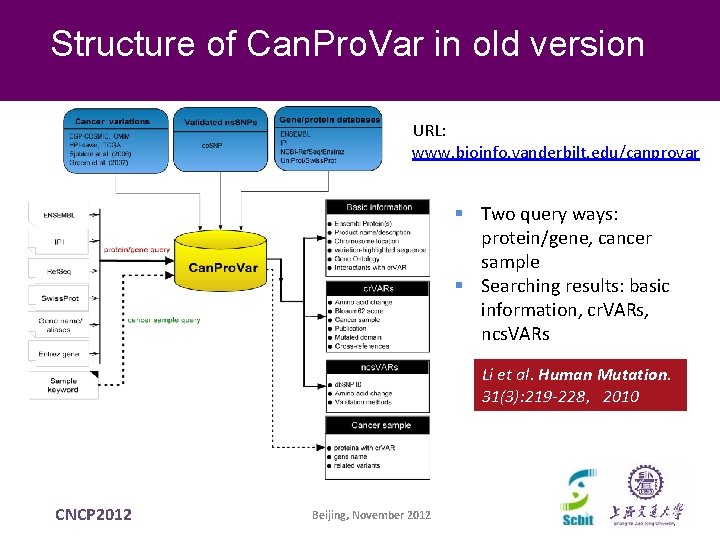
Structure of Can. Pro. Var in old version URL: www. bioinfo. vanderbilt. edu/canprovar § Two query ways: protein/gene, cancer sample § Searching results: basic information, cr. VARs, ncs. VARs Li et al. Human Mutation. 31(3): 219 -228,2010 CNCP 2012 Beijing, November 2012
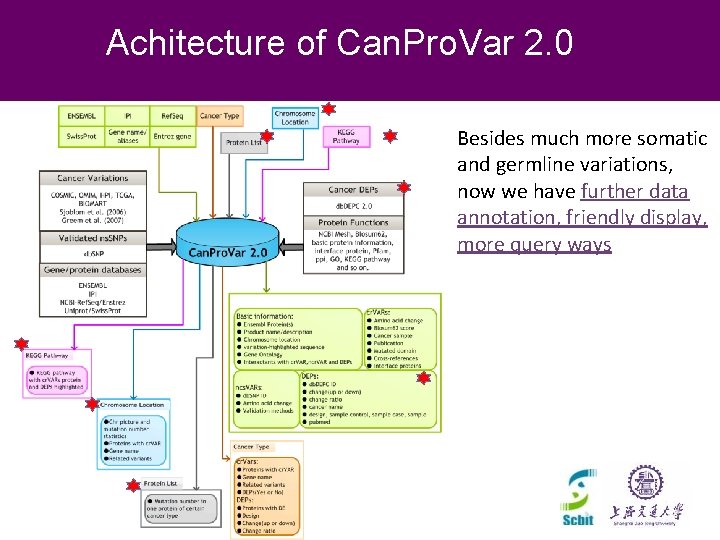
Achitecture of Can. Pro. Var 2. 0 Besides much more somatic and germline variations, now we have further data annotation, friendly display, more query ways CNCP 2012 Beijing, November 2012
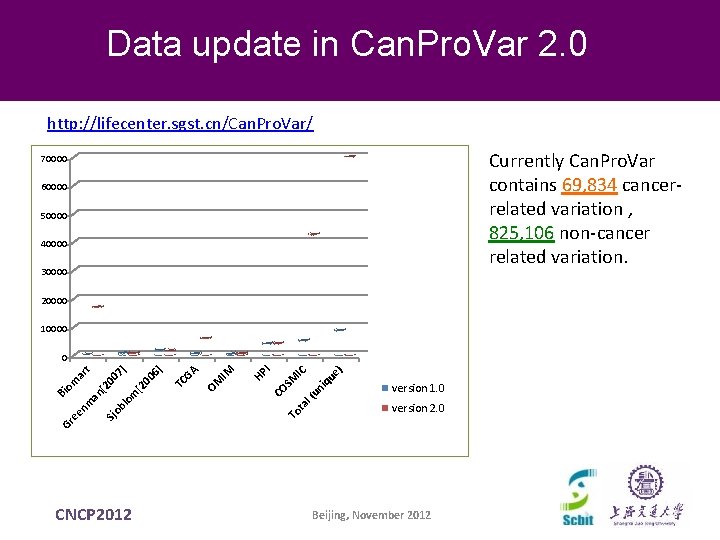
Data update in Can. Pro. Var 2. 0 http: //lifecenter. sgst. cn/Can. Pro. Var/ Currently Can. Pro. Var contains 69, 834 cancerrelated variation , 825, 106 non-cancer related variation. 70000 60000 50000 40000 30000 20000 10000 SM To IC ta l( un iq ue ) I CO HP IM OM GA 00 [2 lo m CNCP 2012 TC 6] 7] 00 [2 an Sj ob Gr ee nm Bi om ar t 0 version 1. 0 version 2. 0 Beijing, November 2012
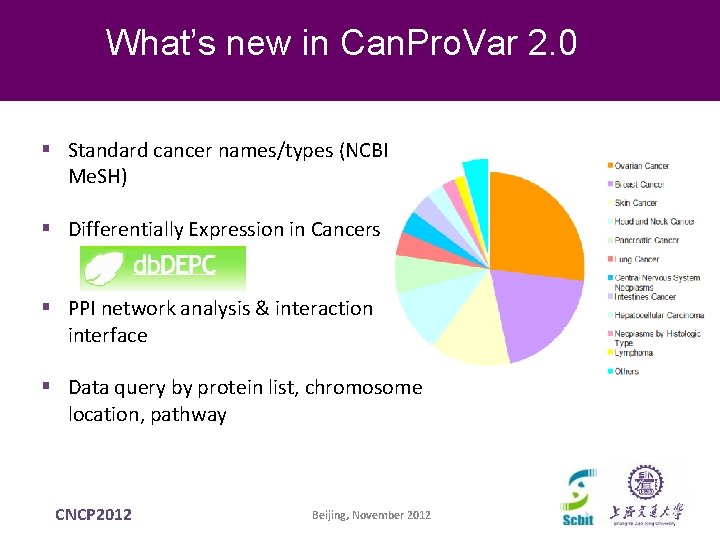
What’s new in Can. Pro. Var 2. 0 § Standard cancer names/types (NCBI Me. SH) § Differentially Expression in Cancers § PPI network analysis & interaction interface § Data query by protein list, chromosome location, pathway CNCP 2012 Beijing, November 2012
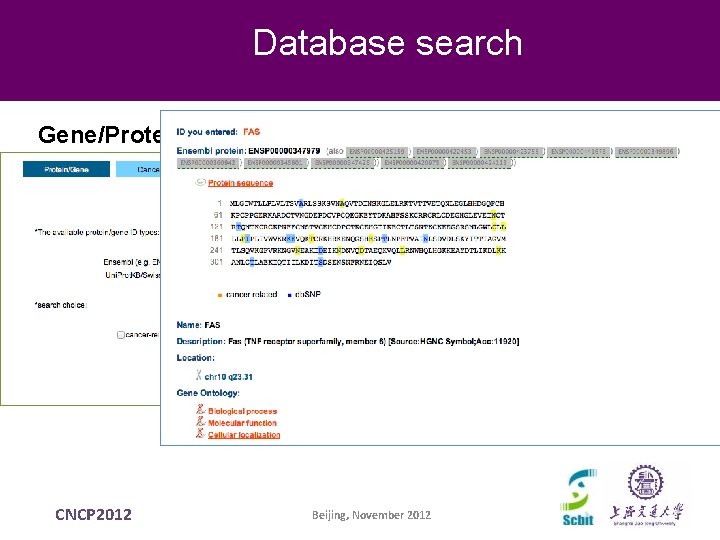
Database search Gene/Protein CNCP 2012 Beijing, November 2012
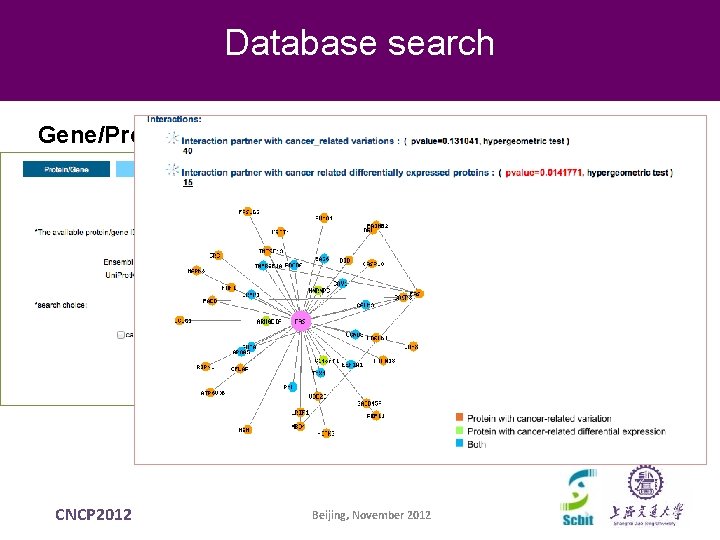
Database search Gene/Protein CNCP 2012 Beijing, November 2012
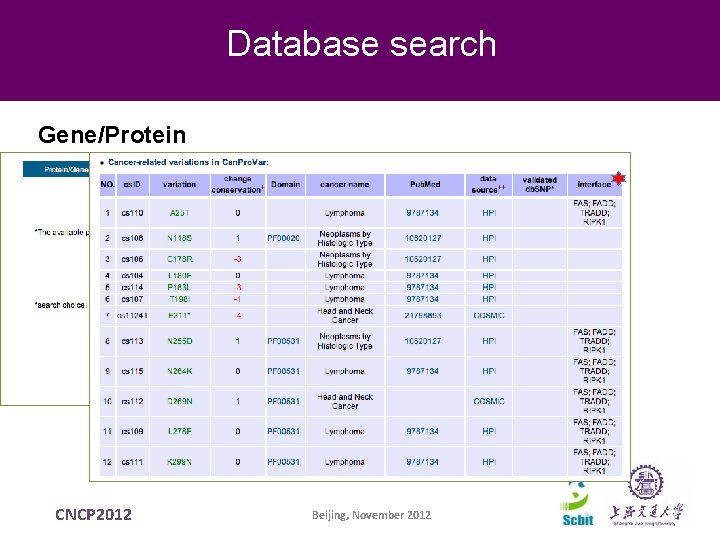
Database search Gene/Protein CNCP 2012 Beijing, November 2012
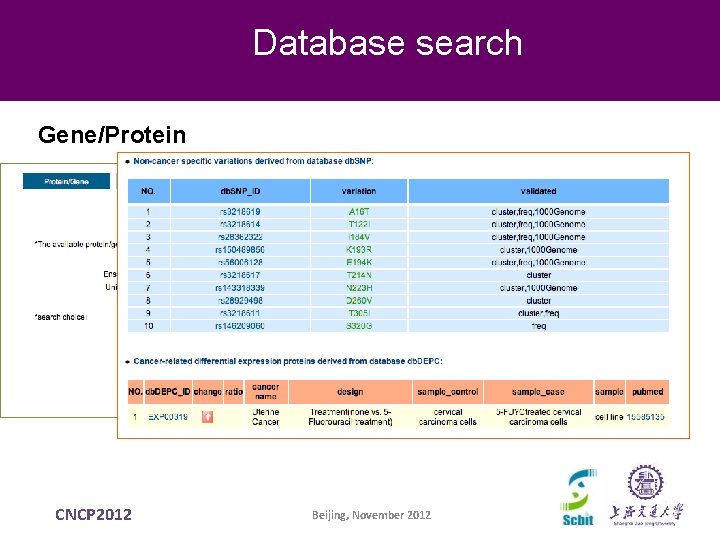
Database search Gene/Protein CNCP 2012 Beijing, November 2012
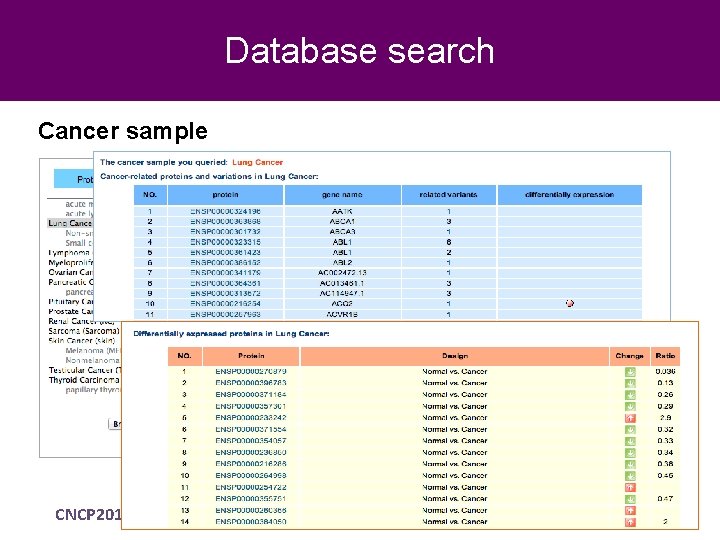
Database search Cancer sample CNCP 2012 Beijing, November 2012
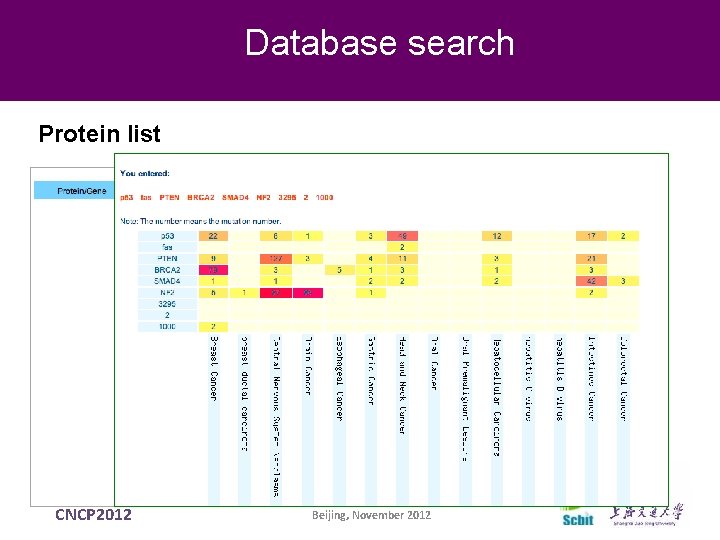
Database search Protein list CNCP 2012 Beijing, November 2012
Database search Chromosome location CNCP 2012 Beijing, November 2012
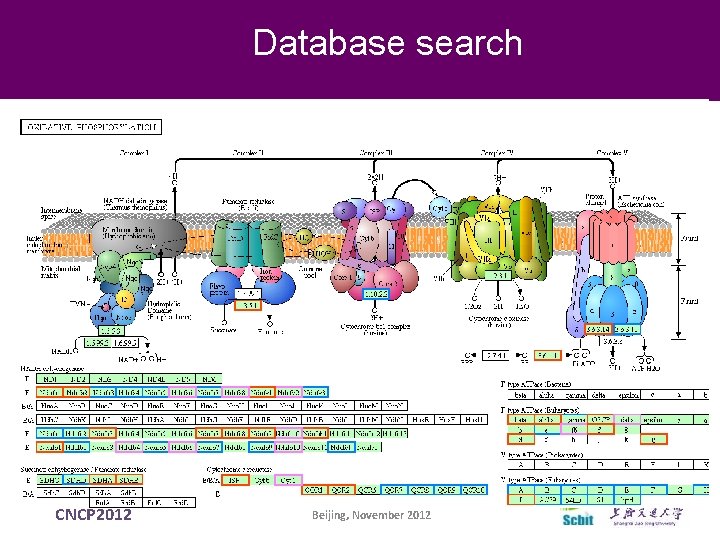
Database search Pathway CNCP 2012 Beijing, November 2012
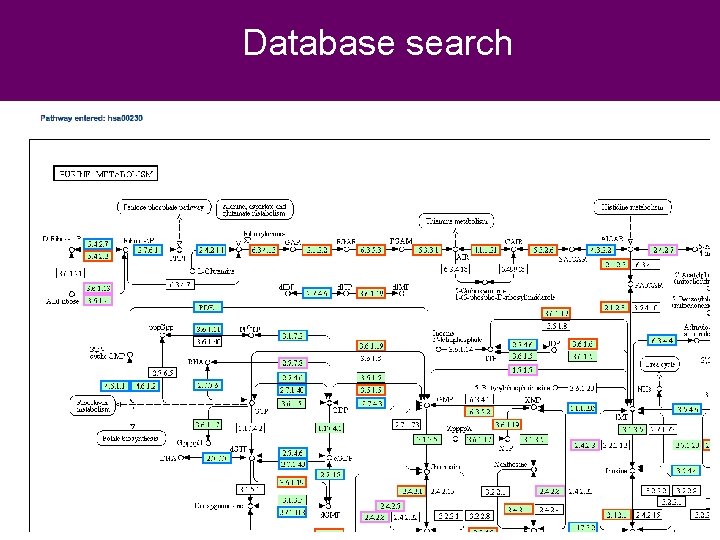
Database search Pathway CNCP 2012 Beijing, November 2012
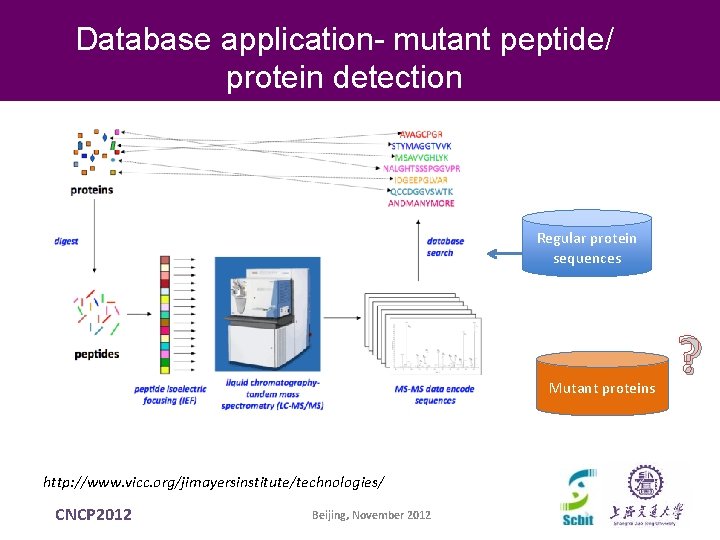
Database application- mutant peptide/ protein detection Regular protein sequences Mutant proteins http: //www. vicc. org/jimayersinstitute/technologies/ CNCP 2012 Beijing, November 2012 ?
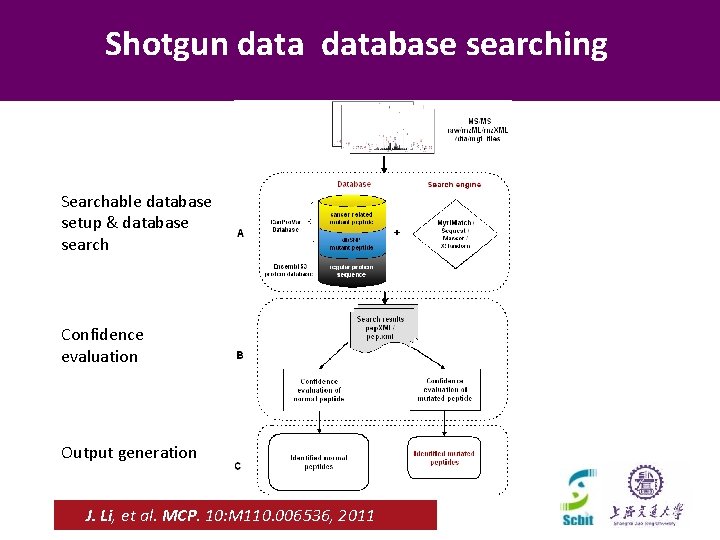
Shotgun database searching Searchable database setup & database search Confidence evaluation Output generation Beijing, November 2012 CNCP 2012 J. Li, et al. MCP. 10: M 110. 006536, 2011
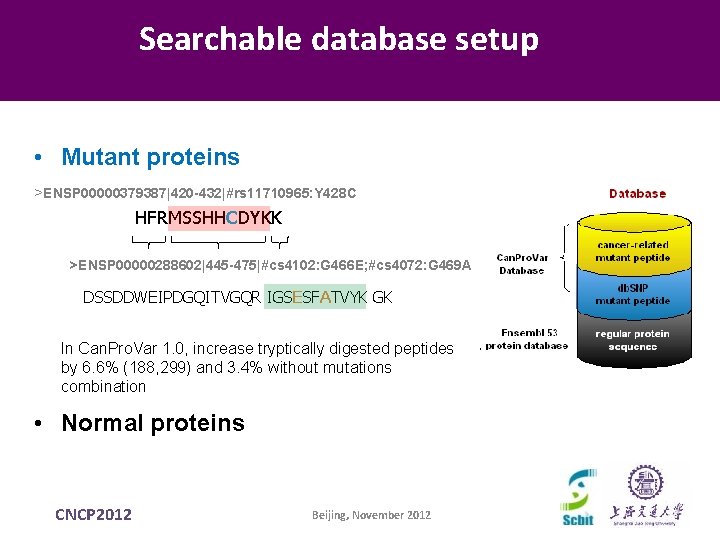
Searchable database setup • Mutant proteins >ENSP 00000379387|420 -432|#rs 11710965: Y 428 C HFRMSSHHCDYKK >ENSP 00000288602|445 -475|#cs 4102: G 466 E; #cs 4072: G 469 A DSSDDWEIPDGQITVGQR IGSESFATVYK GK In Can. Pro. Var 1. 0, increase tryptically digested peptides by 6. 6% (188, 299) and 3. 4% without mutations combination • Normal proteins CNCP 2012 Beijing, November 2012
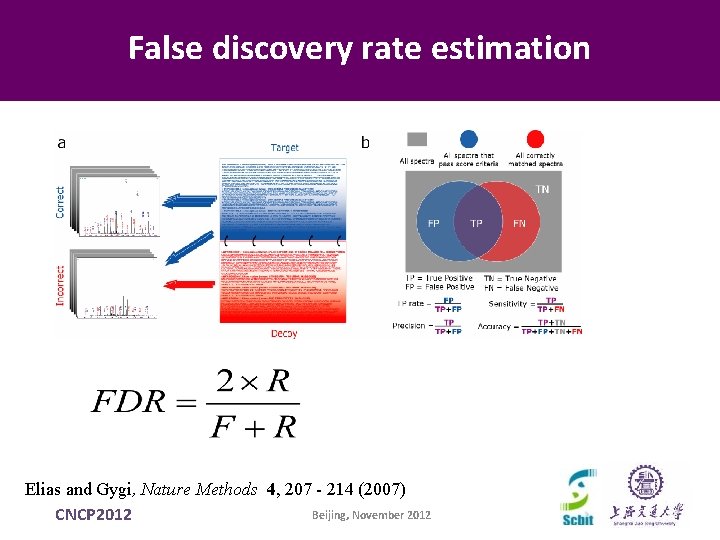
False discovery rate estimation Elias and Gygi, Nature Methods 4, 207 - 214 (2007) Beijing, November 2012 CNCP 2012
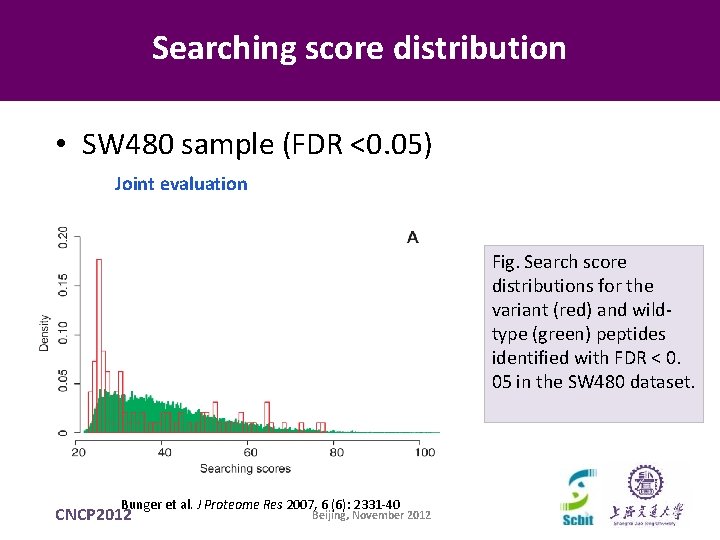
Searching score distribution • SW 480 sample (FDR <0. 05) Joint evaluation Fig. Search score distributions for the variant (red) and wildtype (green) peptides identified with FDR < 0. 05 in the SW 480 dataset. Bunger et al. J Proteome Res 2007, 6 (6): 2331 -40 CNCP 2012 Beijing, November 2012
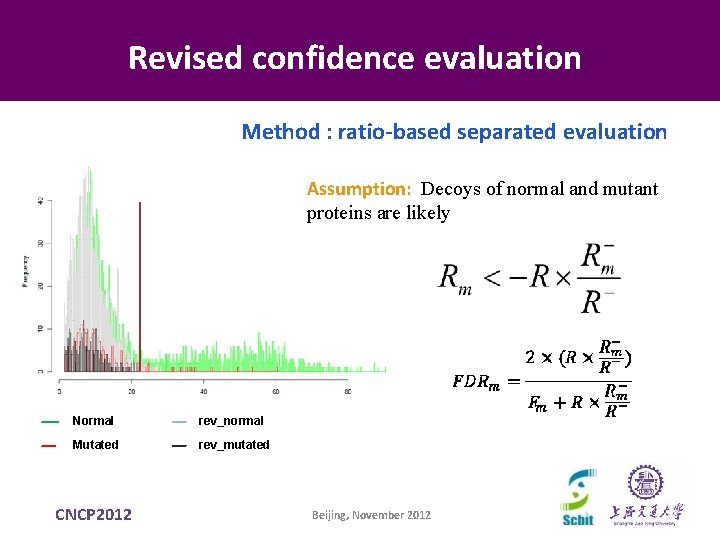
Revised confidence evaluation Method : ratio-based separated evaluation Assumption: Decoys of normal and mutant proteins are likely Normal rev_normal Mutated rev_mutated CNCP 2012 Beijing, November 2012
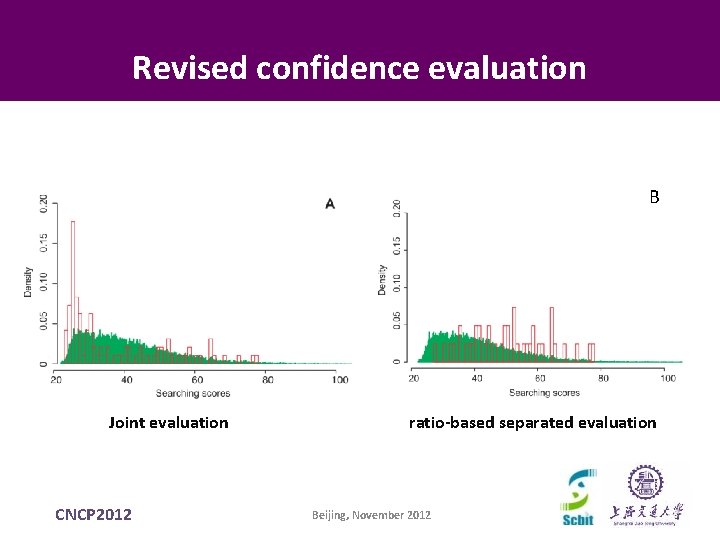
Revised confidence evaluation B Joint evaluation CNCP 2012 ratio-based separated evaluation Beijing, November 2012
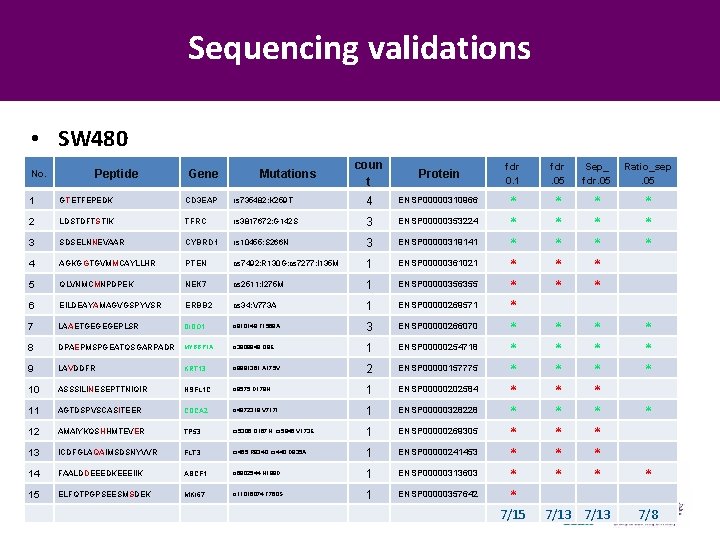
Sequencing validations • SW 480 No. Peptide Gene Mutations coun t Protein 1 GTETFEPEDK CD 3 EAP rs 735482: K 259 T 4 ENSP 00000310966 2 LDSTDFTSTIK TFRC rs 3817672: G 142 S 3 ENSP 00000353224 3 SDSELNNEVAAR CYBRD 1 rs 10455: S 266 N 3 ENSP 00000319141 4 AGKGGTGVMMCAYLLHR PTEN cs 7492: R 130 G; cs 7277: I 135 M 1 ENSP 00000361021 5 QLVNMCMNPDPEK NEK 7 cs 2511: I 275 M 1 ENSP 00000356355 6 EILDEAYAMAGVGSPYVSR ERBB 2 cs 34: V 773 A 1 ENSP 00000269571 7 LAAETGEGEGEPLSR DIDO 1 rs 910148: T 1568 A 3 ENSP 00000266070 8 DPAEPMSPGEATQSGARPADR MYBBP 1 A rs 3809849: Q 8 E 1 ENSP 00000254718 9 LAVDDFR KRT 13 rs 9891361: A 175 V 2 ENSP 00000157775 10 ASSSILINESEPTTNIQIR NSFL 1 C rs 9575: D 179 N 1 ENSP 00000202584 11 AGTDSPVSCASITEER CDCA 2 rs 4872318: V 717 I 1 ENSP 00000328228 12 AMAIYKQSHHMTEVER TP 53 cs 5306: Q 167 H; cs 5945: V 173 E 1 ENSP 00000269305 13 ICDFGLAQAIMSDSNYVVR FLT 3 cs 455: R 834 Q; cs 440: D 835 A 1 ENSP 00000241453 14 FAALDDEEEDKEEEIIK ABCF 1 rs 6902544: N 198 D 1 ENSP 00000313603 15 ELFQTPGPSEESMSDEK MKI 67 rs 11016074: T 760 S 1 ENSP 00000357642 CNCP 2012 Beijing, November 2012 fdr 0. 1 fdr. 05 Sep_ fdr. 05 Ratio_sep. 05 * * * * * * * 7/15 * * * * 7/13 * * * 7/8
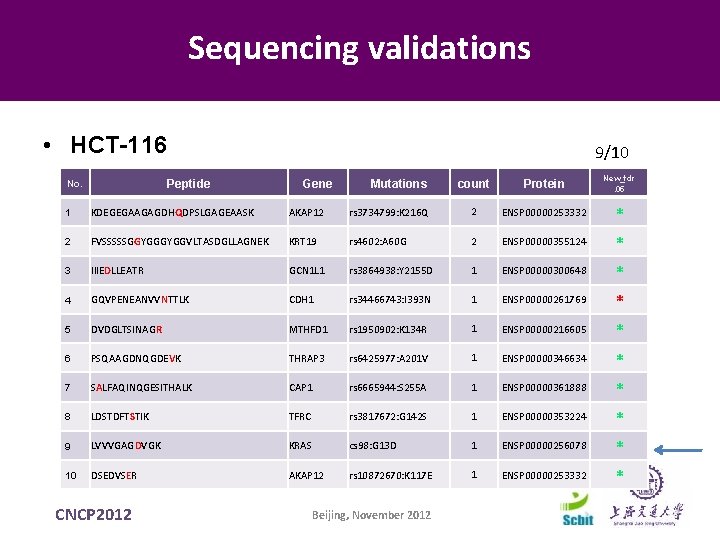
Sequencing validations • HCT-116 Peptide No. 9/10 Gene Mutations count Protein New_fdr. 05 1 KDEGEGAAGAGDHQDPSLGAGEAASK AKAP 12 rs 3734799: K 216 Q 2 ENSP 00000253332 * 2 FVSSSSSGGYGGVLTASDGLLAGNEK KRT 19 rs 4602: A 60 G 2 ENSP 00000355124 * 3 IIIEDLLEATR GCN 1 L 1 rs 3864938: Y 2155 D 1 ENSP 00000300648 * 4 GQVPENEANVVNTTLK CDH 1 rs 34466743: I 393 N 1 ENSP 00000261769 * 5 DVDGLTSINAGR MTHFD 1 rs 1950902: K 134 R 1 ENSP 00000216605 * 6 PSQAAGDNQGDEVK THRAP 3 rs 6425977: A 201 V 1 ENSP 00000346634 * 7 SALFAQINQGESITHALK CAP 1 rs 6665944: S 255 A 1 ENSP 00000361888 * 8 LDSTDFTSTIK TFRC rs 3817672: G 142 S 1 ENSP 00000353224 * 9 LVVVGAGDVGK KRAS cs 98: G 13 D 1 ENSP 00000256078 * 10 DSEDVSER AKAP 12 rs 10872670: K 117 E 1 ENSP 00000253332 * CNCP 2012 Beijing, November 2012
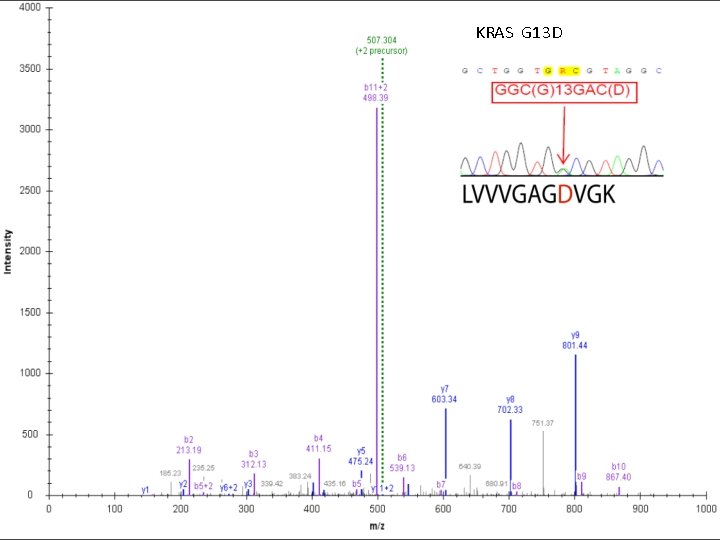
KRAS G 13 D CNCP 2012 Beijing, November 2012 Lab meeting 2/26/2009
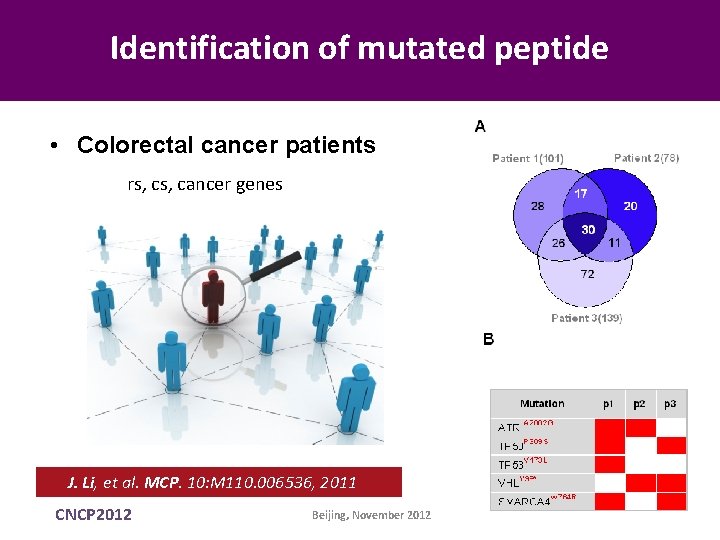
Identification of mutated peptide • Colorectal cancer patients rs, cancer genes J. Li, et al. MCP. 10: M 110. 006536, 2011 CNCP 2012 Beijing, November 2012
Ongoing and future works ² Further mining and test of new Can. Pro. Var data ² Identify p. QTL by protein expression profiles using shotgun proteomics ² Prioritize gene/mutation by integrating sequencing, m. RNA/protein expression and biological networks. CNCP 2012 Beijing, November 2012

Acknowledgements Vanderbilt University SIBS & SCBIT n n Yixue Li Ph. D. Lu Xie Ph. D. n Quoqing Zhang Ph. D. n n n Bing Zhang Ph. D David Tabb Ph. D. Daniel Liebler Ph. D William Pao Ph. D. Zengliu Su Ph. D. SJTU n Menghuan Zhang Ph. D. student n Jia Xu M. Sc. student n Qing Wang M. Sc. student CNCP 2012 Beijing, November 2012

Thank you CNCP 2012 Beijing, November 2012
- Slides: 32