Clustering with kmeans faster smarter cheaper Charles Elkan

























- Slides: 38

Clustering with k-means: faster, smarter, cheaper Charles Elkan University of California, San Diego April 24, 2004

Acknowledgments 1. Funding from Sun Microsystems, with sponsor Dr. Kenny Gross. 2. Advice from colleagues and students, especially Sanjoy Dasgupta (UCSD), Greg Hamerly (Baylor University starting Fall ‘ 04), Doug Turnbull.

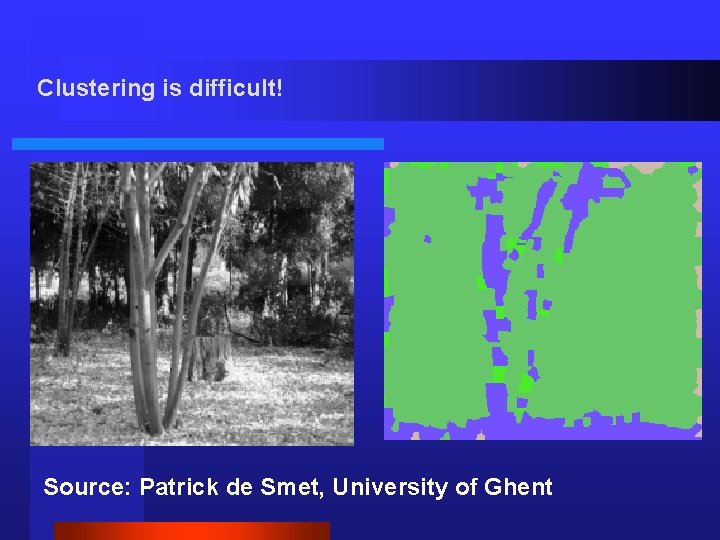
Clustering is difficult! Source: Patrick de Smet, University of Ghent

The standard k-means algorithm Input: n points, distance function d(), number k of clusters to find. STEP NAME Start with k centers 2. Compute d(each point x, each center c) 3. For each x, find closest center c(x) 1. “ALLOCATE” If no point has changed “owner” c(x), stop 5. Each c mean of points owned by it “LOCATE” 6. Repeat from 2 4.

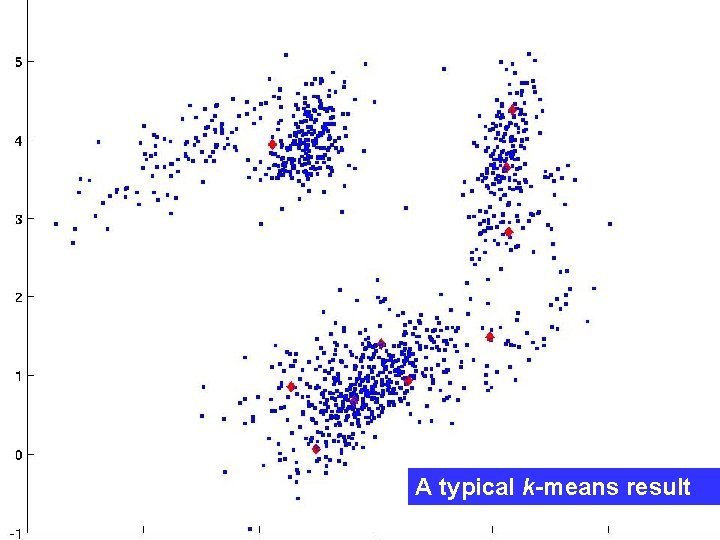
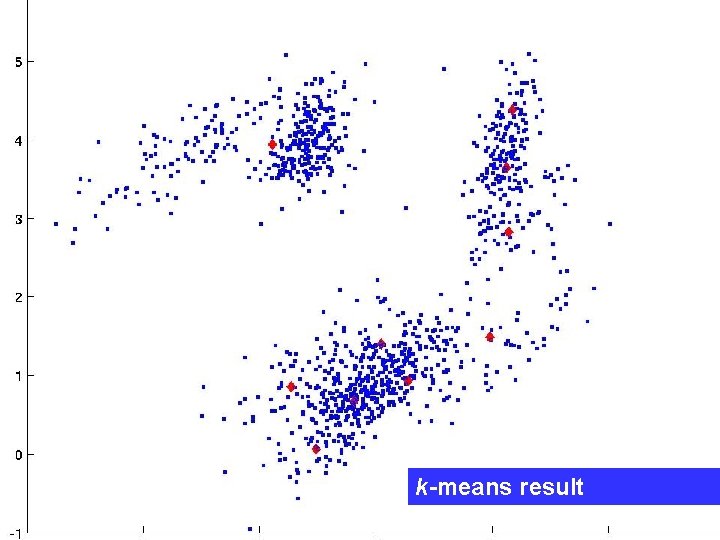
A typical k-means result

Observations Theorem: If d() is Euclidean, then k-means converges monotonically to a local minimum of within-class squared distortion: x d(c(x), x)2 Many variants, complex history since 1956, over 100 papers per year currently 2. Iterative, related to expectation-maximization (EM) 3. # of iterations to converge grows slowly with n, k, d 4. No accepted method exists to discover k. 1.

We want to … … make the algorithm faster. 2. … find lower-cost local minima. (Finding the global optimum is NP-hard. ) 3. … choose the correct k intelligently. 1. With success at (1), we can try more alternatives for (2). With success at (2), comparisons for different k are less likely to be misleading.

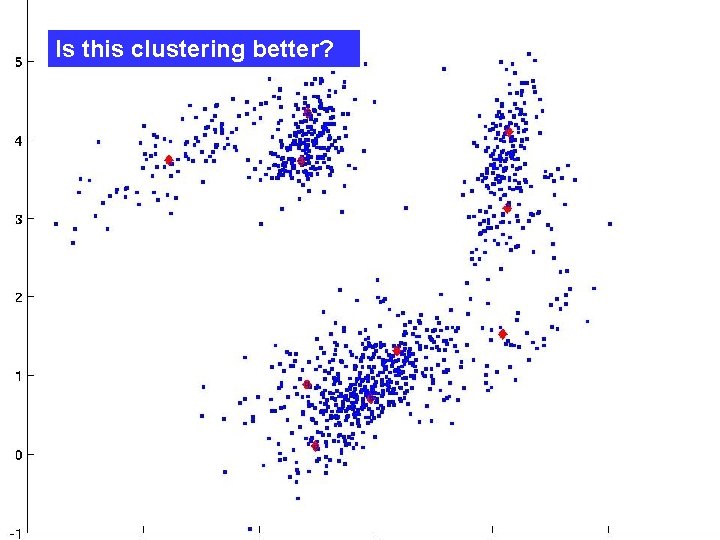
Is this clustering better?

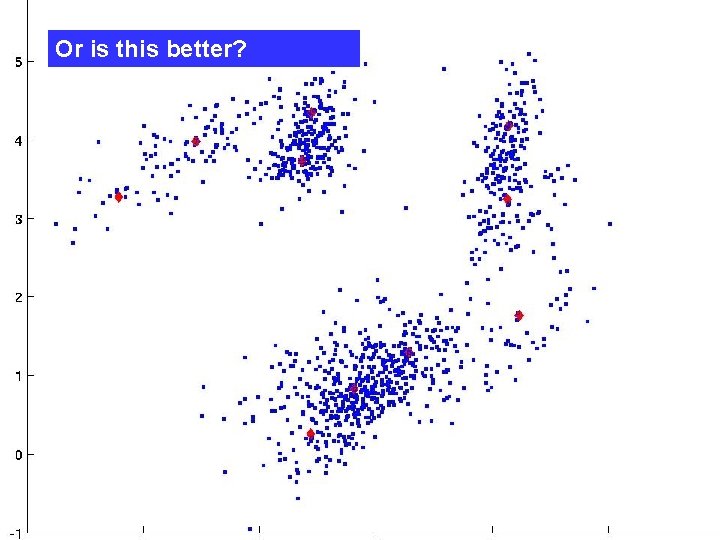
Or is this better?

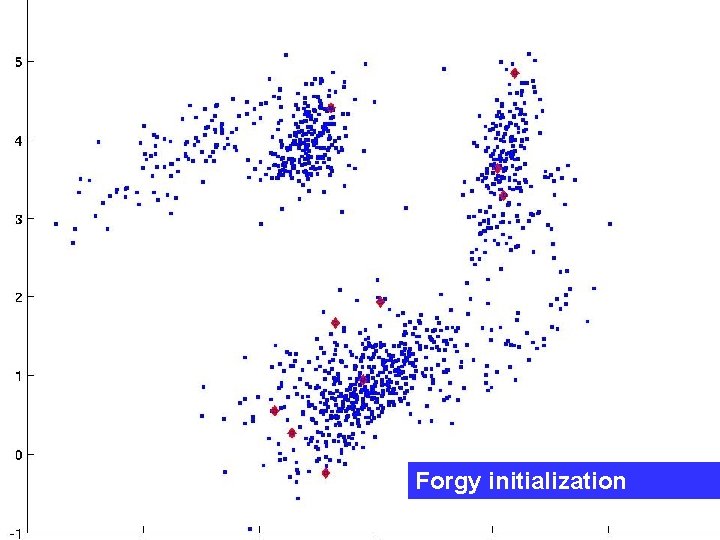
Standard initialization methods Forgy initialization: choose k points at random as starting center locations. Random partitions: divide the data points randomly into k subsets. Both these methods are bad. E. Forgy. Cluster analysis of multivariate data: Efficiency vs. interpretability of classifications. Biometrics, 21(3): 768, 1965.

Forgy initialization

k-means result

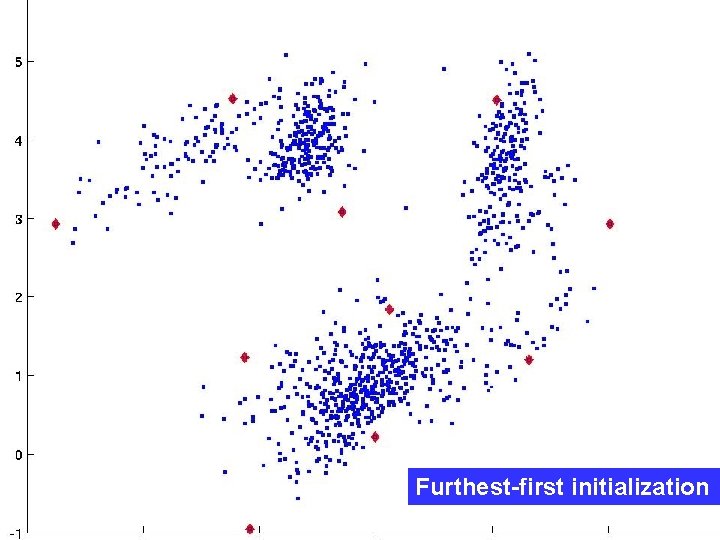
Smarter initialization The “furthest first" algorithm (FF): Pick first center randomly. 2. Next is the point furthest from the first center. 3. Third is the point furthest from both previous centers. 4. In general: next center is argmaxx minc d(x, c) 1. D. Hochbaum, D. Shmoys. A best possible heuristic for the k-center problem, Mathematics of Operations Research, 10(2): 180 -184, 1985.

Furthest-first initialization FF Furthest-first initialization

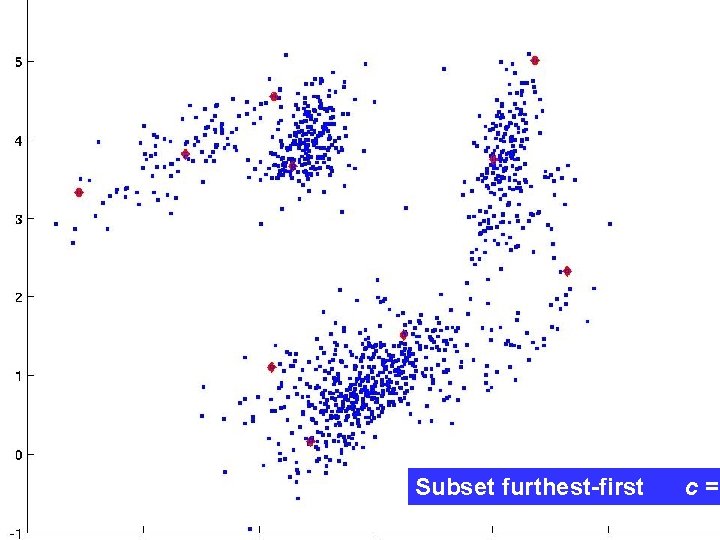
Subset furthest-first (SFF) FF finds outliers, by definition not good cluster centers! Can we choose points far apart and typical of the dataset? Idea: A random sample includes many representative points, but few outliers. But: How big should the random sample be? Lemma: Given k equal-size sets and c >1, with high probability ck log k random points intersect each set.

Subset furthest-first c =

Comparing initialization methods method mean std. dev. best worst Forgy 218 193 29 2201 Furthest-first Subset furthest-first 247 83 59 30 139 20 426 214 218 means 218% worse than the best clustering known. Lower is better.

How to find lower-cost local minima Random restarts, even initialized well, are inadequate. The “central limit catastrophe: ” almost all local minima are only averagely good. K. D. Boese, A. B. Kahng, & S. Muddu, A new adaptive multi-start technique for combinatorial global optimizations. Operations Research Letters 16 (1994) 101 -113. The art of designing a local search algorithm: defining a neighborhood rich in improving candidate moves.

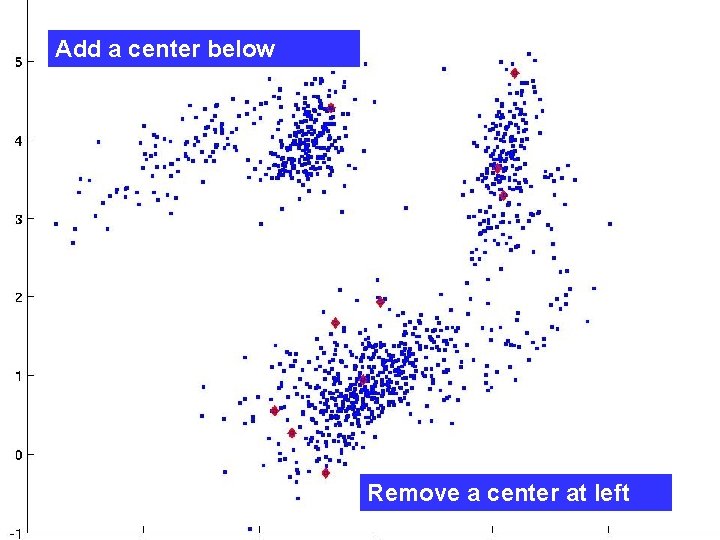
Our local search method k-means alternates two guaranteed-improvement steps: “allocate” and “locate. ” Sadly, we know no other guaranteed-improvement steps. So, we do non-guaranteed “jump” operations: delete an existing center and create a new center at a data point. After each “jump”, run k-means to convergence starting with an “allocate” step.

Add a center below Remove a center at left

Theory versus practice Theorem: Let C be a set of centers such that no “jump” operation improves the value of C. Then C is at most 25 times worse than the global optimum. T. Kanungo et al. A local search approximation algorithm for clustering, ACM Symposium on Computational Geometry, 2002. Our aim: Find heuristics to identify “jump” steps that are likely to be good. Experiments indicate we can solve problems with up to 2000 points and 20 centers optimally.

An upper bound. . . Lemma 1: The maximum loss from removing center c. Proof: l Suppose b is the center closest to c; let B and C be the subsets owned by b and c, with m = |B| and n = |C|. l If B and C merge, the new center is b’ = (mb+nc)/(m+n). l Because c is the mean of C, for any z x in C d(z, x)2 = x in C d(c, x)2 + nd(z, c)2. l So the loss from the merge is nd(b’, c)2 + md(b’, b)2. This computation is cheap, so we do it for every center.

… and a lower bound Suppose we add a new center at point z. Lemma 2: The gain from adding a center at z is at least { x : d(x, c(x)) > d(x, z) } d(x, c(x))2 - d(x, z)2. This computation is more expensive, so we do it for only 2 k log k random candidates z.

Sometimes a jump should only be a jiggle How to use Lemmas 1 and 2: delete the center with smallest maximum loss, make new center at point with greatest minimum gain. This procedure identifies good global improvements. Small-scale improvements come from “jiggling” the center of an existing cluster: moving the center to a point inside the same cluster.

jj-means: the smarter k-means algorithm Run k-means with SFF initialization. 2. Repeat 1. While improvement do Try the best jump according to Lemmas 1 and 2 2. Until improvement do Try a random jiggle 1. “Try” means run k-means to convergence after. l Insert random jumps to satisfy theorem. l

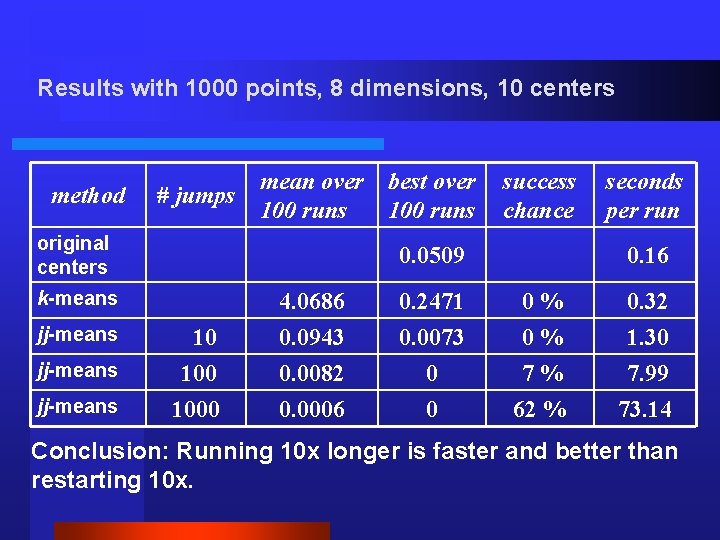
Results with 1000 points, 8 dimensions, 10 centers method # jumps mean over 100 runs original centers jj-means success chance 0. 0509 k-means jj-means best over 100 runs 10 1000 seconds per run 0. 16 4. 0686 0. 2471 0% 0. 32 0. 0943 0. 0082 0. 0006 0. 0073 0 0 0% 7% 62 % 1. 30 7. 99 73. 14 Conclusion: Running 10 x longer is faster and better than restarting 10 x.

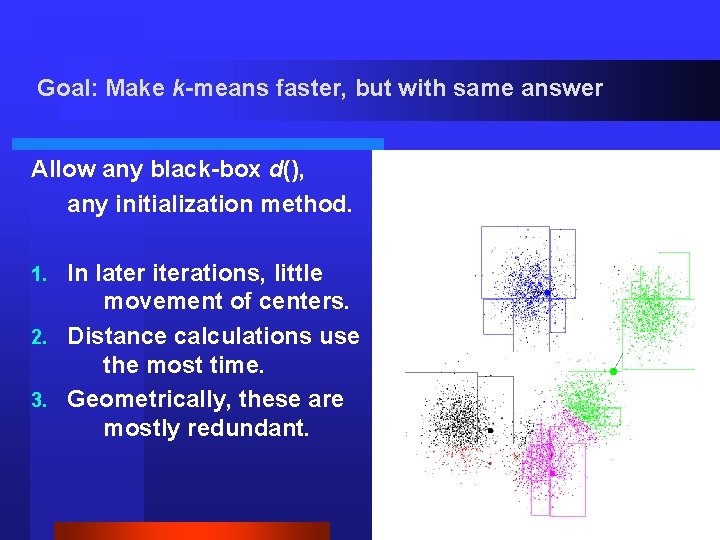
Goal: Make k-means faster, but with same answer Allow any black-box d(), any initialization method. In later iterations, little movement of centers. 2. Distance calculations use the most time. 3. Geometrically, these are mostly redundant. 1. Source: D. Pelleg.

Let x be a point, c(x) its owner, and c a different center. If we already know d(x, c) d(x, c(x)) then computing d(x, c) precisely is not necessary. Strategy: Use the triangle inequality d(x, z) d(x, y) + d(y, z) to get sufficient conditions for d(x, c) d(x, b). kd-trees are useful up to 10 dimensions. l Distance-based data structures can be better. l Our approach is adaptive. l

Lemma 1: Let x be a point, and let b and c be centers. If d(b, c) 2 d(x, b) then d(x, c) d(x, b). Proof: We know d(b, c) d(b, x) + d(x, c). So d(b, c) - d(x, b) d(x, c). Now d(b, c) - d(x, b) 2 d(x, b) - d(x, b) = d(x, b). So d(x, b) d(x, c). • c • x • b

Lemma 2: Let x be a point, let b and c be centers. Then d(x, c) max [ 0, d(x, b) - d(b, c) ]. Proof: We know So Also d(x, b) d(x, c) + d(b, c), d(x, c) d(x, b) - d(b, c). d(x, c) 0. • x • c • b

How to use Lemma 1 Let c(x) be the owner of point x, c' another center: compute d(x, c') only if d(x, c(x)) > ½ d(c(x), c'). If we know an upper bound u(x) d(x, c(x)): compute d(x, c') and d(x, c(x)) only if u(x) > ½ d(c(x), c'). If u(x) ½ min c' c(x) [ d(c(x), c') ]: eliminate all distance calculations for x.

How to use Lemma 2 Let x be any point, let c be any center, let c’ be c at previous iteration. Assume previous lower bound: d(x, c’) l'. Then we get a new lower bound for the current iteration: d(x, c) max [ 0, d(x, c) - d(c, c’)] max [ 0, l' - d(c, c’) ] If l' is a good approximation, and the center only moves slightly, then we get a good updated approximation.

Pick initial centers c. 2. For all x and c, compute d(x, c) 1. Initialize lower bounds l(x, c) d(x, c) 2. Initialize upper bounds u(x) minc d(x, c) 3. Initialize ownership c(x) argminc d(x, c) 3. Repeat until convergence: 1. Find all x s. t. u(x) ½ minc' c(x) [ d(c(x), c') ] 2. For each remaining x and c c(x) s. t. l u(x) > l(x, c) 3. 4. 5. 6. l u(x) > ½ d(c(x), c) 1. Compute d(x, c) and d(x, c(x)) 2. If d(x, c) < d(x, c(x)) then change owner c(x) c 3. Update l(x, c) d(x, c) and u(x) d(x, c(x)) For each c, m(c) mean of points owned by c For each x and c, update l(x, c) max [ 0, l(x, c) - d(m(c), c) ] For each x, update u(x) + d(c(x), m(c(x)) ) Update each center c m(c)

Notes on the new algorithm 1. Empirical issue: which checks to do in which order. 2. Implement “for each remaining x and c” by looping over c, with vectorized code processing all x together. 3. Or, sequentially scan x and l(x, c) from disk. 4. Obvious initialization computes O(nk) distances. Faster methods give inaccurate l(x, c) and u(x), hence may do more distance calculations later.


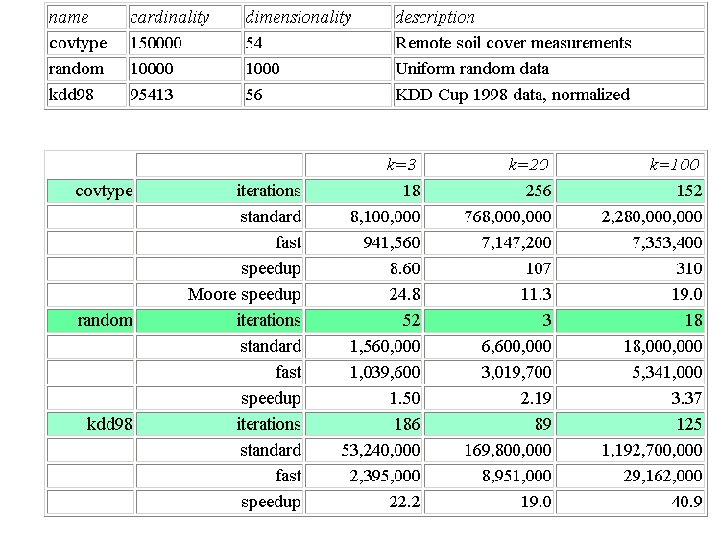
Experimental observations Natural clusters are found while computing the distance between each point and each center less than once! We find k = 100 clusters in n = 100, 000 covtype points with 7, 353, 400 < nk = 15, 000 distance calculations. Number of distance calculations is o(kc), because later iterations compute very few distances.

Current limitations Computing distances is no longer the dominant cost. Reason: After each iteration, we l update nk lower bounds l(x, c) l use O(kd) time to recompute k means l use O(k 2 d) time to recompute all inter-center distances Moreover, we can approximate distances in o(d) time, by considering the largest dimensions first.

Deeper questions What is the minimum # of distance calculations needed? – Adversary argument? If some calculations are omitted, an opponent can choose their values to make any clustering algorithm’s output incorrect. Can we extend to clustering with general Bregman divergences? Can we extend to soft-assignment clustering? Via lower and upper bounds on weights?